5TXE
 
 | |
5TSA
 
 | | Crystal structure of the Zrt-/Irt-like protein from Bordetella bronchiseptica with bound Zn2+ | | Descriptor: | CADMIUM ION, Membrane protein, ZINC ION | | Authors: | Zhang, T, Fellner, M, Sui, D, Liu, J, Hu, J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of a ZIP zinc transporter reveal a binuclear metal center in the transport pathway.
Sci Adv, 3, 2017
|
|
5TXW
 
 | |
5TTD
 
 | | Minor pilin FctB from S. pyogenes with engineered intramolecular isopeptide bond | | Descriptor: | FORMIC ACID, Maltose-binding periplasmic protein,Pilin isopeptide linkage domain protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Young, P.G, Kwon, H, Squire, C.J, Baker, E.N. | | Deposit date: | 2016-11-02 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering a Lys-Asn isopeptide bond into an immunoglobulin-like protein domain enhances its stability.
Sci Rep, 7, 2017
|
|
5TY2
 
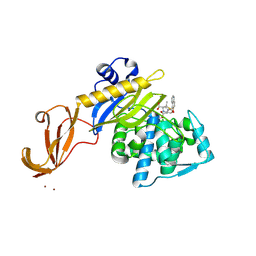 | | Crystal structure of S. aureus penicillin binding protein 4 (PBP4) mutant (E183A, F241R) in complex with nafcillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2-ethoxynaphthalen-1-yl)carbonyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, Penicillin-binding protein 4, ... | | Authors: | Alexander, J.A.N, Strynadka, N.C.J. | | Deposit date: | 2016-11-18 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and kinetic analysis of penicillin-binding protein 4 (PBP4)-mediated antibiotic resistance inStaphylococcus aureus.
J. Biol. Chem., 2018
|
|
5TYI
 
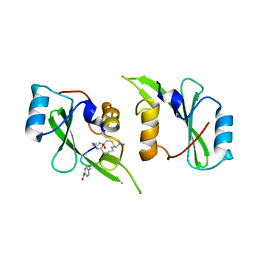 | | Grb7 SH2 with bicyclic peptide containing pY mimetic | | Descriptor: | Growth factor receptor-bound protein 7, Peptide inhibitor | | Authors: | Watson, G.M, Wilce, M.C.J, Wilce, J.A. | | Deposit date: | 2016-11-20 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery, Development, and Cellular Delivery of Potent and Selective Bicyclic Peptide Inhibitors of Grb7 Cancer Target.
J. Med. Chem., 60, 2017
|
|
5TZJ
 
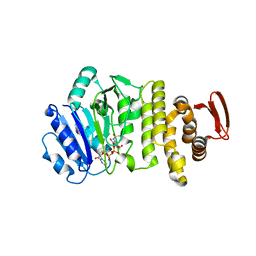 | | Crystal structure of S. aureus TarS 1-349 in complex with UDP-GlcNAc | | Descriptor: | Glycosyl transferase, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Worrall, L.J, Sobhanifar, S, King, D.T, Strynadka, N.C. | | Deposit date: | 2016-11-21 | | Release date: | 2017-01-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Mechanism of Staphylococcus aureus TarS, the Wall Teichoic Acid beta-glycosyltransferase Involved in Methicillin Resistance.
PLoS Pathog., 12, 2016
|
|
5U12
 
 | | E. coli dihydropteroate synthase complexed with an 8-mercaptoguanine derivative: 2-azanyl-8-[(2-fluorophenyl)methylsulfanyl]-1,9-dihydropurin-6-one | | Descriptor: | 2-azanyl-8-[(2-fluorophenyl)methylsulfanyl]-1,9-dihydropurin-6-one, Dihydropteroate synthase | | Authors: | Dennis, M.L, Peat, T.S, Swarbrick, J.D. | | Deposit date: | 2016-11-27 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | 8-Mercaptoguanine Derivatives as Inhibitors of Dihydropteroate Synthase.
Chemistry, 24, 2018
|
|
5U1X
 
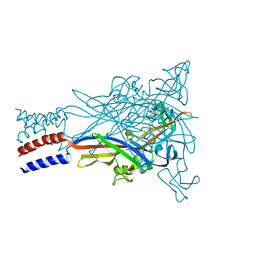 | |
5U6T
 
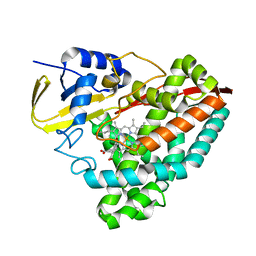 | | The crystal structure of 4-ethoxybenzoate-bound CYP199A4 | | Descriptor: | 4-ethoxybenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Coleman, T, Bruning, J.B, Bell, S.G. | | Deposit date: | 2016-12-08 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Cytochrome P450 CYP199A4 from Rhodopseudomonas palustris Catalyzes Heteroatom Dealkylations, Sulfoxidation, and Amide and Cyclic Hemiacetal Formation
Acs Catalysis, 8, 2018
|
|
5UDD
 
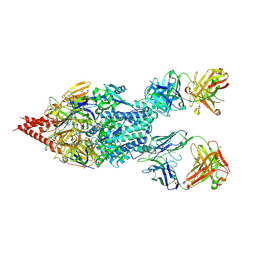 | | Crystal Structure of RSV F B9320 Bound to MEDI8897 | | Descriptor: | Fusion glycoprotein F0, MEDI8897 Fab Heavy Chain, MEDI8897 Fab Light Chain, ... | | Authors: | McLellan, J.S. | | Deposit date: | 2016-12-26 | | Release date: | 2017-05-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants.
Sci Transl Med, 9, 2017
|
|
5UEM
 
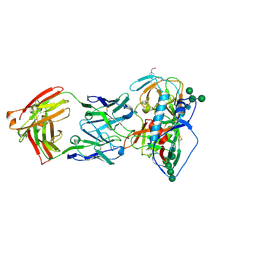 | | Crystal structure of 354NC37 Fab in complex with HIV-1 clade AE strain 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 354NC37 Fab Heavy Chain, ... | | Authors: | Sievers, S.A, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-01-02 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller.
Sci Transl Med, 9, 2017
|
|
5TPQ
 
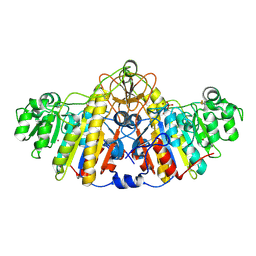 | | E. coli alkaline phosphatase D101A, D153A, R166S, E322A, K328A mutant | | Descriptor: | Alkaline phosphatase, PHOSPHATE ION, ZINC ION | | Authors: | Sunden, F, AlSadhan, I, Lyubimov, A.Y, Doukov, T, Swan, J, Herschlag, D. | | Deposit date: | 2016-10-20 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Differential catalytic promiscuity of the alkaline phosphatase superfamily bimetallo core reveals mechanistic features underlying enzyme evolution.
J. Biol. Chem., 292, 2017
|
|
5TTW
 
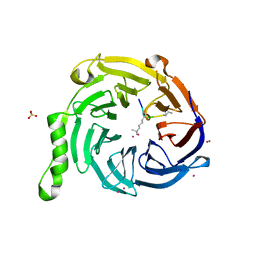 | | Crystal Structure of EED in Complex with UNC4859 | | Descriptor: | Polycomb protein EED, SULFATE ION, UNC4859, ... | | Authors: | The, J, Barnash, K.D, Brown, P.J, Edwards, A.M, Bountra, C, Frye, S.V, James, L.I, Arrowsmith, C.H. | | Deposit date: | 2016-11-04 | | Release date: | 2017-01-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of Peptidomimetic Ligands of EED as Allosteric Inhibitors of PRC2.
ACS Comb Sci, 19, 2017
|
|
5TUQ
 
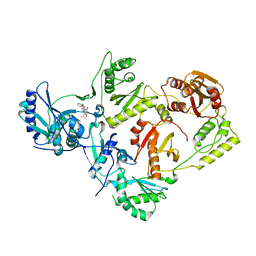 | | Crystal Structure of a 6-Cyclohexylmethyl-3-hydroxypyrimidine-2,4-dione Inhibitor in Complex with HIV Reverse Transcriptase | | Descriptor: | 1-[(benzyloxy)methyl]-6-(cyclohexylmethyl)-3-hydroxy-5-(propan-2-yl)pyrimidine-2,4(1H,3H)-dione, HIV-1 REVERSE TRANSCRIPTASE, MAGNESIUM ION | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2016-11-07 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | 6-Cyclohexylmethyl-3-hydroxypyrimidine-2,4-dione as an inhibitor scaffold of HIV reverase transcriptase: Impacts of the 3-OH on inhibiting RNase H and polymerase.
Eur J Med Chem, 128, 2017
|
|
5TVD
 
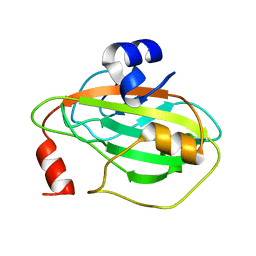 | | Crystal structure of Tm16 | | Descriptor: | Tm16 | | Authors: | Asojo, O.A. | | Deposit date: | 2016-11-08 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.734 Å) | | Cite: | Identification, Characterization, and Structure of Tm16 from Trichuris muris.
J Parasitol Res, 2017, 2017
|
|
5TWZ
 
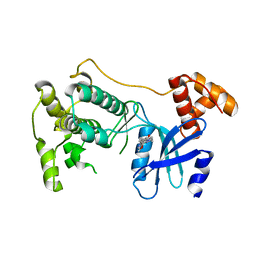 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 7-{[2-methoxy-4-(1H-pyrazol-4-yl)benzoyl]amino}-2,3,4,5-tetrahydro-1H-3-benzazepinium, Maternal embryonic leucine zipper kinase | | Authors: | Seo, H.-Y, Dhe-Paganon, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
5TXY
 
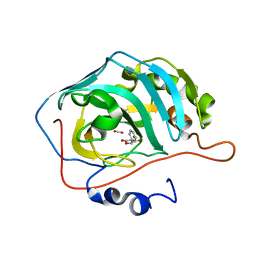 | | Identification of a New Zinc Binding Chemotype of by Fragment Screening on human carbonic anhydrase | | Descriptor: | (5R)-5-phenyl-1,3-oxazolidine-2,4-dione, Carbonic anhydrase 2, FORMIC ACID, ... | | Authors: | Ren, B, Peat, T.S, Poulsen, S.-A. | | Deposit date: | 2016-11-17 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.206 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
5TYA
 
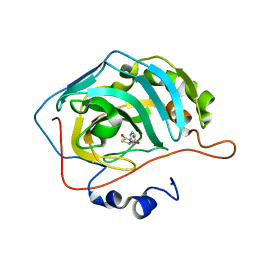 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-phenyl-1,3-thiazolidine-2,4-dione, Carbonic anhydrase 2, ZINC ION | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-18 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
5TZB
 
 | | Burkholderia sp. beta-aminopeptidase | | Descriptor: | CALCIUM ION, D-aminopeptidase | | Authors: | McGowan, S, Drinkwater, N, John, M, Dumsday, G. | | Deposit date: | 2016-11-21 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Crystal structure of a beta-aminopeptidase from an Australian Burkholderia sp.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5U0C
 
 | |
