2ZPH
 
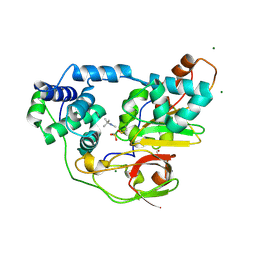 | | Complex of Fe-type nitrile hydratase with tert-butylisonitrile, photo-activated for 340min at 293K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Hashimoto, K, Suzuki, H, Taniguchi, K, Noguchi, T, Yohda, M, Odaka, M. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Catalytic mechanism of nitrile hydratase proposed by time-resolved X-ray crystallography using a novel substrate, tert-butylisonitrile
J.Biol.Chem., 283, 2008
|
|
1WAR
 
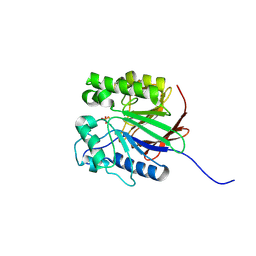 | | Recombinant Human Purple Acid Phosphatase expressed in Pichia Pastoris | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FE (III) ION, HUMAN PURPLE ACID PHOSPHATASE, ... | | Authors: | Duff, A.P, Langley, D.B, Han, R, Averill, B.A, Freeman, H.C, Guss, J.M. | | Deposit date: | 2004-10-28 | | Release date: | 2005-07-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal Structures of Recombinant Human Purple Acid Phosphatase with and without an Inhibitory Conformation of the Repression Loop.
J.Mol.Biol., 351, 2005
|
|
1L4B
 
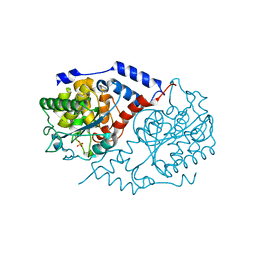 | | Crystal Structure of CobT in apo state | | Descriptor: | Nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase, PHOSPHATE ION | | Authors: | Cheong, C.-G, Escalante-Semerena, J, Rayment, I. | | Deposit date: | 2002-03-04 | | Release date: | 2002-09-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Capture of a labile substrate by expulsion of water molecules from the active site of nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT) from Salmonella enterica.
J.Biol.Chem., 277, 2002
|
|
1A4P
 
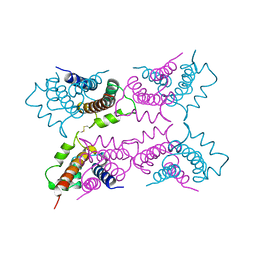 | | P11 (S100A10), LIGAND OF ANNEXIN II | | Descriptor: | S100A10 | | Authors: | Rety, S, Sopkova, J, Renouard, M, Osterloh, D, Gerke, V, Russo-Marie, F, Lewit-Bentley, A. | | Deposit date: | 1998-01-30 | | Release date: | 1998-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure of a complex of p11 with the annexin II N-terminal peptide.
Nat.Struct.Biol., 6, 1999
|
|
6JLV
 
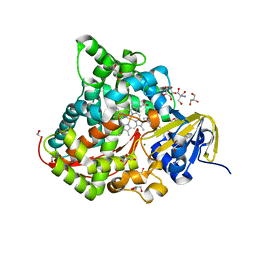 | | Near-Atomic Resolution Structure of the CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Stanfield, J.K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-03-07 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6JMW
 
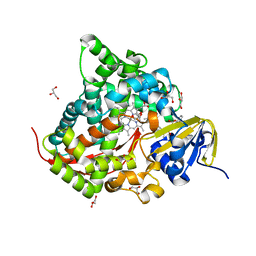 | | Structure of the Chromium Protoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, Chromium Protoporphyrin IX, ... | | Authors: | Stanfield, J.K, Omura, K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-03-13 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
1TC8
 
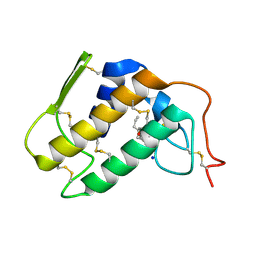 | | Crystal structure of Krait-venom phospholipase A2 in a complex with a natural fatty acid tridecanoic acid | | Descriptor: | N-TRIDECANOIC ACID, SODIUM ION, phospholipase A2 isoform 1 | | Authors: | Singh, G, Jasti, J, Saravanan, K, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | Deposit date: | 2004-05-21 | | Release date: | 2004-06-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the complex formed between a group I phospholipase A2 and a naturally occurring fatty acid at 2.7 A resolution
PROTEIN SCI., 14, 2005
|
|
2ONG
 
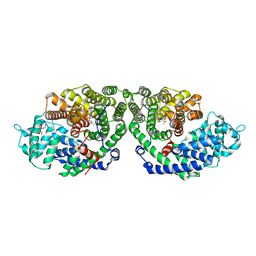 | | Crystal Structure of of limonene synthase with 2-fluorogeranyl diphosphate (FGPP). | | Descriptor: | (1S)-1-[(1R)-1-FLUOROETHYL]-1,5-DIMETHYLHEXYL TRIHYDROGEN DIPHOSPHATE, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4S-limonene synthase, ... | | Authors: | Hyatt, D.C, Youn, B, Croteau, R, Kang, C. | | Deposit date: | 2007-01-23 | | Release date: | 2007-03-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of limonene synthase, a simple model for terpenoid cyclase catalysis.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
6SS9
 
 | | Human Leukocyte Antigen Class I A02 Carrying LLWNGPMHV | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Rizkallah, P.J, Bovay, A. | | Deposit date: | 2019-09-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Identification of a superagonist variant of the immunodominant Yellow fever virus epitope NS4b214-222by combinatorial peptide library screening.
Mol.Immunol., 125, 2020
|
|
5WJO
 
 | | Crystal structure of the unliganded PG90 TCR | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PG90 TCR alpha chain, ... | | Authors: | Shahine, A, Gras, S, Rossjohn, J. | | Deposit date: | 2017-07-24 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A molecular basis of human T cell receptor autoreactivity toward self-phospholipids.
Sci Immunol, 2, 2017
|
|
1RSU
 
 | |
1GDK
 
 | |
7VDN
 
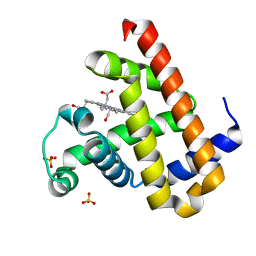 | | High resolution crystal structure of Sperm Whale Myoglobin in the carbonmonoxy form | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shibayama, N, Sato-Tomita, A, Ishimoto, N, Park, S.Y. | | Deposit date: | 2021-09-07 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | X-ray fluorescence holography of biological metal sites: Application to myoglobin.
Biochem.Biophys.Res.Commun., 635, 2022
|
|
2FMG
 
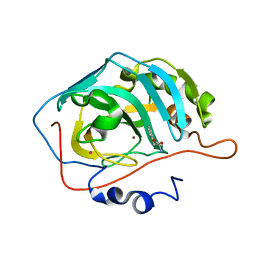 | | Carbonic anhydrase activators. Activation of isoforms I, II, IV, VA, VII and XIV with L- and D- phenylalanine and crystallographic analysis of their adducts with isozyme II: sterospecific recognition within the active site of an enzyme and its consequences for the drug design, structure with L-phenylalanine | | Descriptor: | Carbonic anhydrase 2, MERCURY (II) ION, PHENYLALANINE, ... | | Authors: | Temperini, C, Scozzafava, A, Vullo, D, Supuran, C.T. | | Deposit date: | 2006-01-09 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Carbonic Anhydrase Activators. Activation of Isoforms I, II, IV, VA, VII, and XIV with l- and d-Phenylalanine and Crystallographic Analysis of Their Adducts with Isozyme II: Stereospecific Recognition within the Active Site of an Enzyme and Its Consequences for the Drug Design.
J.Med.Chem., 49, 2006
|
|
2OQJ
 
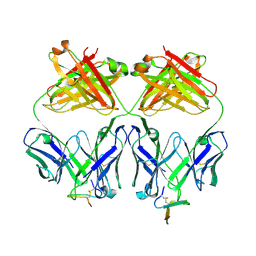 | | Crystal structure analysis of Fab 2G12 in complex with peptide 2G12.1 | | Descriptor: | Fab 2G12 heavy chain, Fab 2G12 light chain, peptide 2G12.1 (ACPPSHVLDMRSGTCLAAEGK) | | Authors: | Calarese, D.A, Stanfield, R.L, Menendez, A, Scott, J.K, Wilson, I.A. | | Deposit date: | 2007-01-31 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A peptide inhibitor of HIV-1 neutralizing antibody 2G12 is not a structural mimic of the natural carbohydrate epitope on gp120.
Faseb J., 22, 2008
|
|
2PO3
 
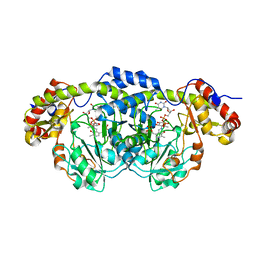 | | Crystal Structure Analysis of DesI in the presence of its TDP-sugar product | | Descriptor: | (2R,3R,4S,5S,6R)-3,4-DIHYDROXY-5-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)IMINO]-6-METHYLTETRAHYDRO-2H-PYRAN-2-YL [(2R,3S,5R)-3-HYDROXY-5-(5-METHYL-2,4-DIOXO-3,4-DIHYDROPYRIMIDIN-1(2H)-YL)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE, 4-dehydrase | | Authors: | Burgie, E.S, Holden, H.M. | | Deposit date: | 2007-04-25 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Architecture of DesI: A Key Enzyme in the Biosynthesis of Desosamine
Biochemistry, 46, 2007
|
|
2POQ
 
 | | Dimeric Dihydrodiol Dehydrogenase complexed with inhibitor, Isoascorbic acid | | Descriptor: | BETA-MERCAPTOETHANOL, Dimeric dihydrodiol dehydrogenase, ISOASCORBIC ACID, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2007-04-27 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structure of monkey dimeric dihydrodiol dehydrogenase in complex with isoascorbic acid.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6GNS
 
 | | Crystal Structure of Leishmania major N-Myristoyltransferase (NMT) With Bound Myristoyl-CoA and an Azepanyl Phenyl Benzylsulphonamide Ligand | | Descriptor: | Glycylpeptide N-tetradecanoyltransferase, TETRADECANOYL-COA, methyl 4-(azepan-1-yl)-3-[[4-[4-(1-methylpiperidin-4-yl)butyl]phenyl]sulfonylamino]benzoate | | Authors: | Robinson, D.A, Harrison, J.R, Brand, S, Smith, V.C, Thompson, S, Smith, A, Davies, K, Mok, N.Y, Torrie, L.S, Collie, I, Hallyburton, I, Norval, S, Simeons, F.R.C, Stojanovski, L, Frearson, J.A, Brenk, R, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2018-05-31 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
J. Med. Chem., 61, 2018
|
|
3DWQ
 
 | | Crystal structure of the A-subunit of the AB5 toxin from E. coli with Neu5Gc-2,3Gal-1,3GlcNAc | | Descriptor: | AZIDE ION, N-PROPANOL, N-glycolyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Byres, E, Paton, A.W, Paton, J.C, Lofling, J.C, Smith, D.F, Wilce, M.C.J, Talbot, U.M, Chong, D.C, Yu, H, Huang, S, Chen, X, Varki, N.M, Varki, A, Rossjohn, J, Beddoe, T. | | Deposit date: | 2008-07-22 | | Release date: | 2008-11-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin
Nature, 456, 2008
|
|
2PTX
 
 | | Crystal Structure of the T. brucei enolase complexed with sulphate in closed conformation | | Descriptor: | 1,2-ETHANEDIOL, Enolase, SULFATE ION, ... | | Authors: | Navarro, M.V.A.S, Rigden, D.J, Garratt, R.C, Dias, S.M.G. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural flexibility in Trypanosoma brucei enolase revealed by X-ray crystallography and molecular dynamics.
Febs J., 274, 2007
|
|
1TOK
 
 | | Maleic acid-bound structure of SRHEPT mutant of E. coli aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, MALEIC ACID | | Authors: | Chow, M.A, McElroy, K.E, Corbett, K.D, Berger, J.M, Kirsch, J.F. | | Deposit date: | 2004-06-14 | | Release date: | 2004-10-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Narrowing substrate specificity in a directly evolved enzyme: the A293D mutant of aspartate aminotransferase
Biochemistry, 43, 2004
|
|
6GJR
 
 | | Cyclophilin A complexed with tri-vector ligand 9. | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, ethyl 2-[[(4-aminophenyl)methyl-[(2-methyl-1,2,3,4-tetrazol-5-yl)methyl]carbamoyl]amino]ethanoate | | Authors: | Georgiou, C, De Simone, A, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-05-16 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A computationally designed binding mode flip leads to a novel class of potent tri-vector cyclophilin inhibitors.
Chem Sci, 10, 2019
|
|
6GJY
 
 | | Cyclophilin A complexed with tri-vector ligand 5. | | Descriptor: | Peptidyl-prolyl cis-trans isomerase A, ethyl 2-[[(4-aminophenyl)methyl-prop-2-ynyl-carbamoyl]amino]ethanoate | | Authors: | Georgiou, C, De Simone, A, Walkinshaw, M.D, Michel, J. | | Deposit date: | 2018-05-17 | | Release date: | 2018-11-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | A computationally designed binding mode flip leads to a novel class of potent tri-vector cyclophilin inhibitors.
Chem Sci, 10, 2019
|
|
2PVR
 
 | | Crystal structure of the catalytic subunit of protein kinase CK2 (C-terminal deletion mutant 1-335) in complex with two sulfate ions | | Descriptor: | Casein kinase II subunit alpha, catalytic subunit, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Niefind, K, Yde, C.W, Ermakova, I, Issinger, O.-G. | | Deposit date: | 2007-05-10 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.605 Å) | | Cite: | Evolved to Be Active: Sulfate Ions Define Substrate Recognition Sites of CK2alpha and Emphasise its Exceptional Role within the CMGC Family of Eukaryotic Protein Kinases
J.Mol.Biol., 370, 2007
|
|
1RXC
 
 | | E. COLI uridine phosphorylase: 5-fluorouracil ribose-1-phosphate complex | | Descriptor: | 1-O-phosphono-alpha-D-ribofuranose, 5-FLUOROURACIL, 5-FLUOROURIDINE, ... | | Authors: | Caradoc-Davies, T.T, Cutfield, S.M, Lamont, I.L, Cutfield, J.F. | | Deposit date: | 2003-12-18 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of escherichia coli uridine phosphorylase in two native and three complexed forms reveal basis of substrate specificity, induced conformational changes and influence of potassium
J.Mol.Biol., 337, 2004
|
|
