7H6H
 
 | | THE 1.94 A CRYSTAL STRUCTURE OF HUMAN CATHEPSIN G IN COMPLEX WITH 1-[(7-fluoronaphthalen-1-yl)methyl]-3-[[methoxycarbonyl(methyl)amino]methyl]indole-2-carboxylic acid | | Descriptor: | 1-[(7-fluoronaphthalen-1-yl)methyl]-3-{[(methoxycarbonyl)(methyl)amino]methyl}-1H-indole-2-carboxylic acid, Cathepsin G, SULFATE ION, ... | | Authors: | Banner, D.W, Benz, J.M, Schlatter, D, Hilpert, H. | | Deposit date: | 2024-04-19 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structures of human Chymase and Cathepsin G
To be published
|
|
5Z8F
 
 | | Solution structure for the unique dimeric 4:2 complex of a platinum(II)-based tripod bound to a hybrid-1 human telomeric G-quadruplex | | Descriptor: | 4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]-N,N-bis[4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]phenyl]aniline, G-quadruplex DNA (26-MER) | | Authors: | Liu, W.T, Zhong, Y.F, Liu, L.Y, Zeng, W.J, Wang, F.Y, Yang, D.Z, Mao, Z.W. | | Deposit date: | 2018-01-31 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding
Nat Commun, 9, 2018
|
|
3UGX
 
 | | Crystal Structure of Visual Arrestin | | Descriptor: | 1,2-ETHANEDIOL, IMIDAZOLE, PENTANEDIAL, ... | | Authors: | Batra-Safferling, R, Granzin, J. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.649 Å) | | Cite: | Crystal Structure of p44, a Constitutively Active Splice Variant of Visual Arrestin.
J.Mol.Biol., 416, 2012
|
|
4E90
 
 | | Human phosphodiesterase 9 in complex with inhibitors | | Descriptor: | 6-[(3S,4S)-4-methyl-1-(pyrimidin-2-ylmethyl)pyrrolidin-3-yl]-1-(tetrahydro-2H-pyran-4-yl)-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Liu, S. | | Deposit date: | 2012-03-20 | | Release date: | 2013-02-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Application of structure-based drug design and parallel chemistry to identify selective, brain penetrant, in vivo active phosphodiesterase 9A inhibitors.
J.Med.Chem., 55, 2012
|
|
2MF8
 
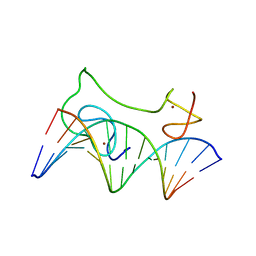 | | HADDOCK model of MyT1 F4F5 - DNA complex | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*AP*AP*AP*GP*TP*TP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*AP*AP*CP*TP*TP*TP*CP*GP*GP*T)-3'), Myelin transcription factor 1, ... | | Authors: | Gamsjaeger, R, O'Connell, M.R, Cubeddu, L, Shepherd, N.E, Lowry, J.A, Kwan, A.H, Vandevenne, M, Swanton, M.K, Matthews, J.M, Mackay, J.P. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A structural analysis of DNA binding by myelin transcription factor 1 double zinc fingers.
J.Biol.Chem., 288, 2013
|
|
3H6W
 
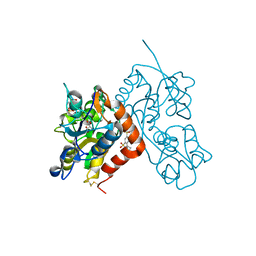 | | Crystal structure of the iGluR2 ligand-binding core (S1S2J-N754S) in complex with glutamate and NS5217 at 1.50 A resolution | | Descriptor: | (3R)-3-cyclopentyl-6-methyl-7-[(4-methylpiperazin-1-yl)sulfonyl]-3,4-dihydro-2H-1,2-benzothiazine 1,1-dioxide, DIMETHYL SULFOXIDE, GLUTAMIC ACID, ... | | Authors: | Hald, H, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2009-04-24 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Distinct structural features of cyclothiazide are responsible for effects on peak current amplitude and desensitization kinetics at iGluR2.
J.Mol.Biol., 391, 2009
|
|
6T2M
 
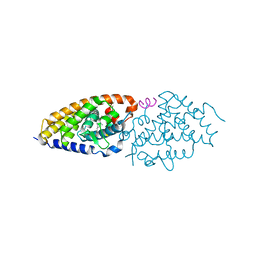 | | VDR-ZK168281 complex | | Descriptor: | Nuclear receptor coactivator 1, Vitamin D3 receptor A, ethyl (~{Z})-3-[1-[(~{E},1~{R},4~{R})-4-[(1~{R},3~{a}~{S},4~{E},7~{a}~{R})-7~{a}-methyl-4-[(2~{Z})-2-[(3~{S},5~{R})-2-methylidene-3,5-bis(oxidanyl)cyclohexylidene]ethylidene]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-1-yl]-1-oxidanyl-pent-2-enyl]cyclopropyl]prop-2-enoate | | Authors: | Rochel, N, Belorusova, A.Y. | | Deposit date: | 2019-10-09 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Analysis of VDR Complex with ZK168281 Antagonist.
J.Med.Chem., 63, 2020
|
|
3UF4
 
 | |
4IMF
 
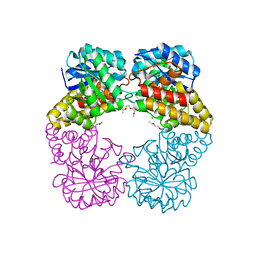 | |
3UG5
 
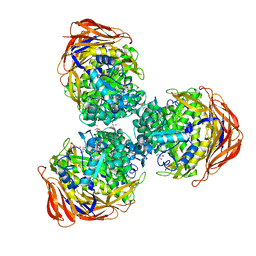 | | Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Alpha-L-arabinofuranosidase, beta-D-xylopyranose | | Authors: | Im, D.-H, Miyazaki, K, Wakagi, T, Fushinobu, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Glycoside Hydrolase Family 51 alpha-L-Arabinofuranosidase from Thermotoga maritima
Biosci.Biotechnol.Biochem., 76, 2012
|
|
3OPZ
 
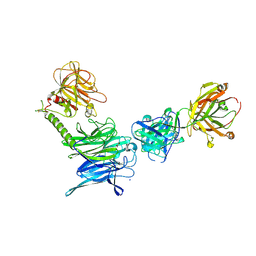 | | Crystal structure of trans-sialidase in complex with the Fab fragment of a neutralizing monoclonal IgG antibody | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, SODIUM ION, Trans-sialidase, ... | | Authors: | Larrieux, N, Muia, R, Campetella, O, Buschiazzo, A. | | Deposit date: | 2010-09-02 | | Release date: | 2011-11-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Trypanosoma cruzi trans-sialidase in complex with a neutralizing antibody: structure/function studies towards the rational design of inhibitors.
Plos Pathog., 8, 2012
|
|
4CDL
 
 | | Crystal Structure of Retro-aldolase RA110.4-6 Complexed with Inhibitor 1-(6-methoxy-2-naphthalenyl)-1,3-butanedione | | Descriptor: | (2E)-1-(6-methoxynaphthalen-2-yl)but-2-en-1-one, STEROID DELTA-ISOMERASE | | Authors: | Pinkas, D.M, Studer, S, Obexer, R, Giger, L, Gruetter, M.G, Baker, D, Hilvert, D. | | Deposit date: | 2013-11-01 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Active Site Plasticity of a Computationally Designed Retro-Aldolase Enzyme
To be Published
|
|
3E8L
 
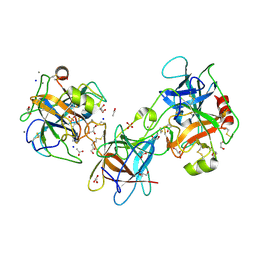 | | The Crystal Structure of the Double-headed Arrowhead Protease Inhibitor A in Complex with Two Trypsins | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Bao, R, Jiang, C.-H, Chi, C.W, Lin, S.X, Chen, Y.X. | | Deposit date: | 2008-08-20 | | Release date: | 2009-07-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The ternary structure of double-headed arrowhead protease inhibitor API-A complexed with two trypsins reveals a novel reactive site conformation.
J.Biol.Chem., 284, 2009
|
|
4OLF
 
 | |
1I1N
 
 | | HUMAN PROTEIN L-ISOASPARTATE O-METHYLTRANSFERASE WITH S-ADENOSYL HOMOCYSTEINE | | Descriptor: | PROTEIN-L-ISOASPARTATE O-METHYLTRANSFERASE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Smith, C.D, Chattopadhyay, D, Carson, M, Friedman, A.M, Skinner, M.M. | | Deposit date: | 2001-02-02 | | Release date: | 2002-03-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human L-isoaspartyl-O-methyl-transferase with S-adenosyl homocysteine at 1.6-A resolution and modeling of an isoaspartyl-containing peptide at the active site.
Protein Sci., 11, 2002
|
|
3EGT
 
 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-722 | | Descriptor: | 1-(2,2-diphosphonoethyl)-3-(heptyloxy)pyridinium, Farnesyl pyrophosphate synthase, MAGNESIUM ION | | Authors: | Cao, R, Gao, Y, Robinson, H, Oldfield, E. | | Deposit date: | 2008-09-11 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation.
J.Am.Chem.Soc., 131, 2009
|
|
2R9B
 
 | | Structural Analysis of Plasmepsin 2 from Plasmodium falciparum complexed with a peptide-based inhibitor | | Descriptor: | Plasmepsin-2, peptide-based inhibitor | | Authors: | Liu, P, Marzahn, M.R, Robbins, A.H, McKenna, R, Dunn, B.M. | | Deposit date: | 2007-09-12 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recombinant plasmepsin 1 from the human malaria parasite plasmodium falciparum: enzymatic characterization, active site inhibitor design, and structural analysis.
Biochemistry, 48, 2009
|
|
3EG6
 
 | |
5XF3
 
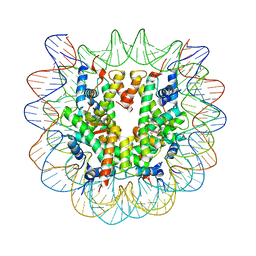 | | Nucleosome core particle with an adduct of a binuclear RAPTA (Ru-arene-phosphaadamantane) compound having a 1,2-diphenylethylenediamine linker (R,R-configuration) | | Descriptor: | (1R,2R)-1,2-diphenylethane-1,2-diamine, DNA (145-MER), Histone H2A type 1-B/E, ... | | Authors: | Ma, Z, Adhireksan, Z, Murray, B.S, Dyson, P.J, Davey, C.A. | | Deposit date: | 2017-04-07 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nucleosome acidic patch-targeting binuclear ruthenium compounds induce aberrant chromatin condensation
Nat Commun, 8, 2017
|
|
4O7C
 
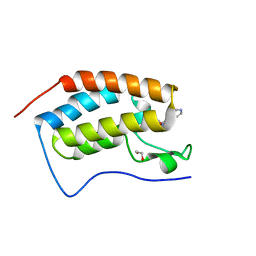 | | Crystal structure of the first bromodomain of human BRD4 in complex with SB-614067-R | | Descriptor: | 1,2-ETHANEDIOL, 4-[(5Z)-5-(1-nitroso-2,3-dihydro-5H-inden-5-ylidene)-2-(piperidin-4-yl)-3,5-dihydro-4H-imidazol-4-ylidene]-1,4-dihydropyridine, Bromodomain-containing protein 4 | | Authors: | Ember, S.W, Zhu, J.-Y, Watts, C, Schonbrunn, E. | | Deposit date: | 2013-12-24 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Acetyl-lysine Binding Site of Bromodomain-Containing Protein 4 (BRD4) Interacts with Diverse Kinase Inhibitors.
Acs Chem.Biol., 9, 2014
|
|
5X8L
 
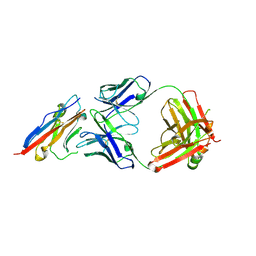 | | PD-L1 in complex with atezolizumab | | Descriptor: | Programmed cell death 1 ligand 1, atezolizumab heavy chain, atezolizumab light chain | | Authors: | Heo, Y.S, Lee, H.T. | | Deposit date: | 2017-03-03 | | Release date: | 2017-08-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular mechanism of PD-1/PD-L1 blockade via anti-PD-L1 antibodies atezolizumab and durvalumab
Sci Rep, 7, 2017
|
|
4O9D
 
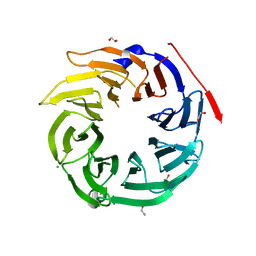 | | Structure of Dos1 propeller | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Rik1-associated factor 1 | | Authors: | Kuscu, C, Schalch, T, Joshua-Tor, L. | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRL4-like Clr4 complex in Schizosaccharomyces pombe depends on an exposed surface of Dos1 for heterochromatin silencing.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5SDT
 
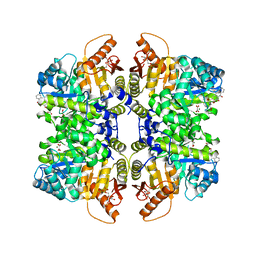 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 15 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-(3,4-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)-beta-alanine, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2022-01-20 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5XAU
 
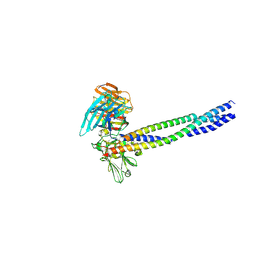 | | Crystal structure of integrin binding fragment of laminin-511 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Takizawa, M, Arimori, T, Kitago, Y, Takagi, J, Sekiguchi, K. | | Deposit date: | 2017-03-15 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanistic basis for the recognition of laminin-511 by alpha 6 beta 1 integrin.
Sci Adv, 3, 2017
|
|
1HGT
 
 | |
