7QEF
 
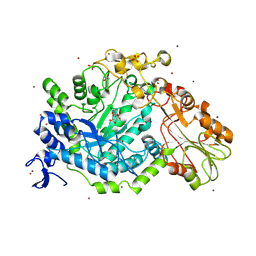 | | Crystal structure of para-nitrophenyl-Beta-D-glucuronide bound to a mutant of SN243 (D415A) | | Descriptor: | 4-nitrophenyl beta-D-glucopyranosiduronic acid, ACETATE ION, SN243, ... | | Authors: | Neun, S, Brear, P, Campbell, E, Omari, K, Wagner, O, Hyvonen, M, Hollfelder, F. | | Deposit date: | 2021-12-02 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Functional metagenomic screening identifies an unexpected beta-glucuronidase.
Nat.Chem.Biol., 18, 2022
|
|
6P5J
 
 | | Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 2) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
7BYR
 
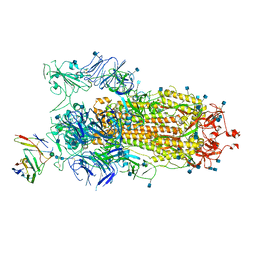 | | BD23-Fab in complex with the S ectodomain trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ab23-Fab-Heavy Chain, ... | | Authors: | Zhu, Q, Wang, G, Xiao, J. | | Deposit date: | 2020-04-24 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients' B Cells.
Cell, 182, 2020
|
|
8TNH
 
 | |
8GN2
 
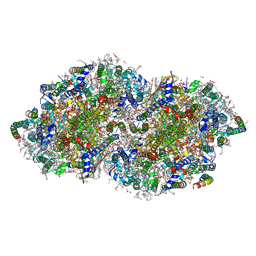 | | Crystal structure of PPBQ-bound photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kamada, S, Nakajima, Y, Shen, J.-R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the action mechanisms of artificial electron acceptors in photosystem II.
J.Biol.Chem., 299, 2023
|
|
7C07
 
 | | Crystal structure of yeast U2AF1 complex bound to 5'-AAGGU RNA. | | Descriptor: | RNA (5'-R(*U*AP*AP*GP*GP*U)-3'), Splicing factor U2AF 23 kDa subunit, Splicing factor U2AF 59 kDa subunit, ... | | Authors: | Yoshida, H, Park, S.Y, Urano, T, Obayashi, E. | | Deposit date: | 2020-04-30 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Elucidation of the aberrant 3' splice site selection by cancer-associated mutations on the U2AF1.
Nat Commun, 11, 2020
|
|
8TNI
 
 | | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing bi-specific antibody CAP256L-R27 targeting the CD4-binding site and the V2-apex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 BG505 DS-SOSIP gp120, ... | | Authors: | Zhou, T, Morano, N.C, Roark, R.S, Kwong, P.D, Xu, J. | | Deposit date: | 2023-08-01 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Cryo-EM structure of HIV-1 Env BG505 DS-SOSIP in complex with broadly neutralizing bi-specific antibody CAP256L-R27 targeting the CD4-binding site and the V2-apex
To Be Published
|
|
7QGG
 
 | | Neuronal RNA granules are ribosome complexes stalled at the pre-translocation state | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Pulk, A, Kipper, K, Mansour, A. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Neuronal RNA granules are ribosome complexes stalled at the pre-translocation state.
J.Mol.Biol., 434, 2022
|
|
8GN0
 
 | | Crystal structure of DCBQ-bound photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kamada, S, Nakajima, Y, Shen, J.-R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into the action mechanisms of artificial electron acceptors in photosystem II.
J.Biol.Chem., 299, 2023
|
|
7BSB
 
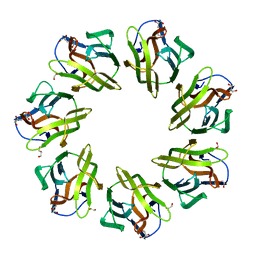 | | Mevo lectin- Native form-2 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, lectin | | Authors: | Sivaji, N, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 2020-03-30 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural and related studies on Mevo lectin from Methanococcus voltae A3: the first thorough characterization of an archeal lectin and its interactions.
Glycobiology, 31, 2021
|
|
7Q3X
 
 | |
6OF1
 
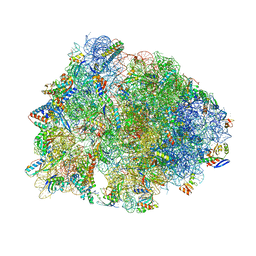 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with dirithromycin and bound to mRNA and A-, P-, and E-site tRNAs at 2.80A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Khabibullina, N.F, Tereshchenkov, A.G, Komarova, E.S, Syroegin, E.A, Shiriaev, D.I, Paleskava, A, Kartsev, V.G, Bogdanov, A.A, Konevega, A.L, Dontsova, O.A, Sergiev, P.V, Osterman, I.A, Polikanov, Y.S. | | Deposit date: | 2019-03-28 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Dirithromycin Bound to the Bacterial Ribosome Suggests New Ways for Rational Improvement of Macrolides.
Antimicrob.Agents Chemother., 63, 2019
|
|
8GJV
 
 | |
8GTP
 
 | | cryo-EM structure of Omicron BA.5 S protein in complex with XGv289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
6OL7
 
 | |
7QK5
 
 | |
8RHL
 
 | | Yeast 20S proteasome in complex with a linear biarylether epoxyketone (compound 15a) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Linear biarylether epoxyketone, ... | | Authors: | Goetz, M.G, Godwin, K, Price, R, Dorn, R, Merrill-Steskal, G, Hansen, H, Klemmer, W, Produturi, G, Rocha, M, Palmer, M, Molacek, L, Strater, Z, Groll, M. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Macrocyclic Oxindole Peptide Epoxyketones-A Comparative Study of Macrocyclic Inhibitors of the 20S Proteasome.
Acs Med.Chem.Lett., 15, 2024
|
|
6OM2
 
 | |
8G7L
 
 | | ATP-bound mtHsp60 V72I | | Descriptor: | 60 kDa heat shock protein, mitochondrial, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8RHK
 
 | | Yeast 20S proteasome in complex with a linear oxindole epoxyketone (compound 6) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Linear oxindole epoxyketone, ... | | Authors: | Goetz, M.G, Godwin, K, Price, R, Dorn, R, Merrill-Steskal, G, Hansen, H, Klemmer, W, Produturi, G, Rocha, M, Palmer, M, Molacek, L, Strater, Z, Groll, M. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Macrocyclic Oxindole Peptide Epoxyketones-A Comparative Study of Macrocyclic Inhibitors of the 20S Proteasome.
Acs Med.Chem.Lett., 15, 2024
|
|
8R0B
 
 | | Cryo-EM structure of the cross-exon pre-B+ATP complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
7QSD
 
 | | Bovine complex I in the active state at 3.1 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Bridges, H.R, Blaza, J.N, Yin, Z, Chung, I, Hirst, J. | | Deposit date: | 2022-01-13 | | Release date: | 2022-03-02 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of mammalian respiratory complex I inhibition by medicinal biguanides.
Science, 379, 2023
|
|
8GN1
 
 | | Crystal structure of DBBQ-bound photosystem II complex | | Descriptor: | (3R)-beta,beta-caroten-3-ol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Kamada, S, Nakajima, Y, Shen, J.-R. | | Deposit date: | 2022-08-22 | | Release date: | 2023-07-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the action mechanisms of artificial electron acceptors in photosystem II.
J.Biol.Chem., 299, 2023
|
|
8GTO
 
 | | cryo-EM structure of Omicron BA.5 S protein in complex with XGv282 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Xia, X.Y, Zhang, Y.Y, Chi, X.M, Huang, B.D, Wu, L.S, Zhou, Q. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Comprehensive structural analysis reveals broad-spectrum neutralizing antibodies against SARS-CoV-2 Omicron variants.
Cell Discov, 9, 2023
|
|
8G7N
 
 | | ATP- and mtHsp10-bound mtHsp60 V72I | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
