3HO8
 
 | |
6R01
 
 | | Streptomyces lividans Ccsp mutant - H107A/H111A | | Descriptor: | COPPER (I) ION, Cytosolic copper storage protein | | Authors: | Straw, M.L, Hough, M.A, Worrall, J.A. | | Deposit date: | 2019-03-12 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | A Histidine Residue and a Tetranuclear Cuprous-thiolate Cluster Dominate the Copper Loading Landscape of a Copper Storage Protein from Streptomyces lividans.
Chemistry, 25, 2019
|
|
3HBL
 
 | | Crystal Structure of S. aureus Pyruvate Carboxylase T908A Mutant | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Tong, L, Yu, L.P.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | A Symmetrical Tetramer for S. aureus Pyruvate Carboxylase in Complex with Coenzyme A.
Structure, 17, 2009
|
|
6V1X
 
 | | Cryo-EM Structure of the Hyperpolarization-Activated Potassium Channel KAT1: Tetramer | | Descriptor: | (2S)-1-(nonanoyloxy)-3-(phosphonooxy)propan-2-yl tetradecanoate, (3beta,5beta,14beta,17alpha)-cholestan-3-ol, Potassium channel KAT1 | | Authors: | Clark, M.D, Contreras, G.F, Shen, R, Perozo, E. | | Deposit date: | 2019-11-21 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Electromechanical coupling in the hyperpolarization-activated K + channel KAT1.
Nature, 583, 2020
|
|
3H4R
 
 | | Crystal structure of E. coli RecE exonuclease | | Descriptor: | Exodeoxyribonuclease 8 | | Authors: | Bell, C.E, Zhang, J. | | Deposit date: | 2009-04-20 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of E. coli RecE protein reveals a toroidal tetramer for processing double-stranded DNA breaks.
Structure, 17, 2009
|
|
3O2M
 
 | |
3FDC
 
 | | Crystal Structure of Avidin | | Descriptor: | Avidin, iron(II) tetracyano-5-(2-Oxo-hexahydro-thieno[3,4-d]imidazol-6-yl)-pentanoic acid (4'-methyl-[2,2']bipyridinyl-4-ylmethyl)-amide | | Authors: | Barker, K.D, Sazinsky, M.H, Eckermann, A.L, Abajian, C, Hartings, M.R, Rosenzweig, A.C, Meade, T.J. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Protein Binding and the Electronic Properties of Iron(II) Complexes: An Electrochemical and Optical Investigation of Outer Sphere Effects.
Bioconjug.Chem., 20, 2009
|
|
4Q9N
 
 | | Crystal structure of Chlamydia trachomatis enoyl-ACP reductase (FabI) in complex with NADH and AFN-1252 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide | | Authors: | Yao, J, Abdelrahman, Y, Robertson, R.M, Cox, J.V, Belland, R.J, White, S.W, Rock, C.O. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Type II Fatty Acid Synthesis Is Essential for the Replication of Chlamydia trachomatis.
J.Biol.Chem., 289, 2014
|
|
6B6U
 
 | | Pyruvate Kinase M2 mutant - S437Y | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Srivastava, D, Dey, M. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Investigation of a Dimeric Variant of Pyruvate Kinase Muscle Isoform 2.
Biochemistry, 56, 2017
|
|
4D8Z
 
 | | Crystal structure of B. anthracis DHPS with compound 24 | | Descriptor: | (3R)-3-(7-amino-4,5-dioxo-1,4,5,6-tetrahydropyrimido[4,5-c]pyridazin-3-yl)butanoic acid, Dihydropteroate Synthase, SULFATE ION | | Authors: | Hammoudeh, D, Lee, R.E, White, S.W. | | Deposit date: | 2012-01-11 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structure-Based Design of Novel Pyrimido[4,5-c]pyridazine Derivatives as Dihydropteroate Synthase Inhibitors with Increased Affinity.
Chemmedchem, 7, 2012
|
|
2PES
 
 | | Urate Oxidase in complex with tris-dipicolinate Lutetium | | Descriptor: | 8-AZAXANTHINE, LUTETIUM (III) ION, PYRIDINE-2,6-DICARBOXYLIC ACID, ... | | Authors: | Pompidor, G, Vicat, J, Kahn, R. | | Deposit date: | 2007-04-03 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A dipicolinate lanthanide complex for solving protein structures using anomalous diffraction.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
8UP6
 
 | | Structure of atypical asparaginase from Rhodospirillum rubrum (mutant K19A) in complex with L-Asp | | Descriptor: | ASPARTIC ACID, Asparaginase, TETRAETHYLENE GLYCOL | | Authors: | Lubkowski, J, Wlodawer, A, Zhang, D. | | Deposit date: | 2023-10-21 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | RrA, an enzyme from Rhodospirillum rubrum, is a prototype of a new family of short-chain L-asparaginases.
Protein Sci., 33, 2024
|
|
4D9P
 
 | | Crystal structure of B. anthracis DHPS with compound 17 | | Descriptor: | (3R)-3-(7-amino-1-methyl-4,5-dioxo-1,4,5,6-tetrahydropyrimido[4,5-c]pyridazin-3-yl)butanoic acid, Dihydropteroate Synthase, SULFATE ION | | Authors: | Hammoudeh, D, Lee, R.E, White, S.W. | | Deposit date: | 2012-01-11 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure-Based Design of Novel Pyrimido[4,5-c]pyridazine Derivatives as Dihydropteroate Synthase Inhibitors with Increased Affinity.
Chemmedchem, 7, 2012
|
|
4N3E
 
 | | Crystal structure of Hyp-1, a St John's wort PR-10 protein, in complex with 8-anilino-1-naphthalene sulfonate (ANS) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-ANILINO-1-NAPHTHALENE SULFONATE, Phenolic oxidative coupling protein, ... | | Authors: | Sliwiak, J, Dauter, Z, Mccoy, A.J, Read, R.J, Jaskolski, M. | | Deposit date: | 2013-10-07 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Likelihood-based molecular-replacement solution for a highly pathological crystal with tetartohedral twinning and sevenfold translational noncrystallographic symmetry.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4D8A
 
 | | Crystal structure of B. anthracis DHPS with compound 21 | | Descriptor: | Dihydropteroate synthase, LYSINE, SULFATE ION, ... | | Authors: | Hammoudeh, D, Lee, R.E, White, S.W. | | Deposit date: | 2012-01-10 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.183 Å) | | Cite: | Structure-Based Design of Novel Pyrimido[4,5-c]pyridazine Derivatives as Dihydropteroate Synthase Inhibitors with Increased Affinity.
Chemmedchem, 7, 2012
|
|
4E44
 
 | |
4BX5
 
 | | cis-divalent streptavidin | | Descriptor: | 1,2-ETHANEDIOL, STREPTAVIDIN, TETRAETHYLENE GLYCOL | | Authors: | Fairhead, M, Krndija, D, Lowe, E.D, Howarth, M. | | Deposit date: | 2013-07-08 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.431 Å) | | Cite: | Plug-and-Play Pairing Via Defined Divalent Streptavidins.
J.Mol.Biol., 426, 2014
|
|
3HB9
 
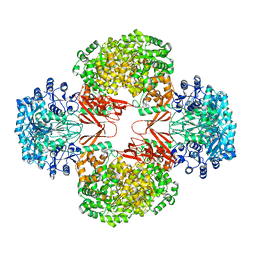 | | Crystal Structure of S. aureus Pyruvate Carboxylase A610T Mutant | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Tong, L, Yu, L.P.C. | | Deposit date: | 2009-05-04 | | Release date: | 2009-06-30 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Symmetrical Tetramer for S. aureus Pyruvate Carboxylase in Complex with Coenzyme A.
Structure, 17, 2009
|
|
4BO7
 
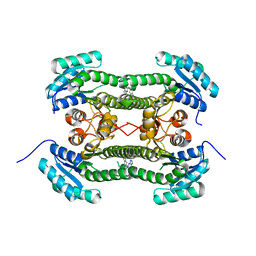 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with N-(2,3-dihydro-1H-inden- 5-yl)tetrazolo(1,5-b)pyridazin-6-amine at 2.6A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, N-(2,3-dihydro-1H-inden-5-yl)tetrazolo[1,5-b]pyridazin-6-amine | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
4ZZ7
 
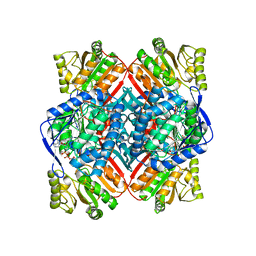 | | Crystal structure of methylmalonate-semialdehyde dehydrogenase (DddC) from Oceanimonas doudoroffii | | Descriptor: | Methylmalonate-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Do, H, Lee, C.W, Lee, S.G, Kang, H, Park, C.M, Kim, H.J, Park, H, Park, H, Lee, J.H. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure and modeling of the tetrahedral intermediate state of methylmalonate-semialdehyde dehydrogenase (MMSDH) from Oceanimonas doudoroffii.
J. Microbiol., 54, 2016
|
|
3RQX
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis in complex with P1,P4-Di(adenosine-5') tetraphosphate | | Descriptor: | ADP/ATP-DEPENDENT NAD(P)H-HYDRATE DEHYDRATASE, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, CHLORIDE ION, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
1ZNA
 
 | |
3KL3
 
 | | Crystal structure of Ligand bound XynC | | Descriptor: | D-HISTIDINE, Glucuronoxylanase xynC, TETRAETHYLENE GLYCOL, ... | | Authors: | St John, F.J, Hurlbert, J.C, Pozharski, E. | | Deposit date: | 2009-11-06 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Ligand bound structures of a glycosyl hydrolase family 30 glucuronoxylan xylanohydrolase.
J.Mol.Biol., 407, 2011
|
|
4UWG
 
 | | Discovery of (2S)-8-((3R)-3-Methylmorpholin-4-yl)-1-(3-methyl-2-oxo- butyl)-2-(trifluoromethyl)-3,4-dihydro-2H-pyrimido(1,2-a)pyrimidin-6- one: a Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors | | Descriptor: | (8S)-2-(morpholin-4-yl)-9-[2-(propan-2-yloxy)ethyl]-8-(trifluoromethyl)-6,7,8,9-tetrahydro-4H-pyrimido[1,2-a]pyrimidin-4-one, PHOSPHATIDYLINOSITOL 3-KINASE CATALYTIC SUBUNIT TYPE 3, SULFATE ION | | Authors: | Pasquier, B, El-Ahmad, Y, Filoche-Romme, B, Dureuil, C, Fassy, F, Abecassis, P.Y, Mathieu, M, Bertrand, T, Benard, T, Barriere, C, ElBatti, S, Letallec, J.P, Sonnefraud, V, Brollo, M, Delbarre, L, Loyau, V, Pilorge, F, Bertin, L, Richepin, P, Arigon, J, Labrosse, J.R, Clement, J, Durand, F, Combet, R, Perraut, P, Leroy, V, Gay, F, Lefrancois, D, Bretin, F, Marquette, J.P, Michot, N, Caron, A, Castell, C, Schio, L, McCort, G, Goulaouic, H, Garcia-Echeverria, C, Ronan, B. | | Deposit date: | 2014-08-12 | | Release date: | 2014-11-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-Yl]-1-(3-Methyl-2-Oxobutyl)-2-(Trifluoromethyl)-3,4-Dihydro-2H-Pyrimido[1,2-A]Pyrimidin-6-One: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors.
J.Med.Chem., 58, 2015
|
|
1I3E
 
 | | HUMAN AZIDO-MET HEMOGLOBIN BART'S (GAMMA4) | | Descriptor: | AZIDE ION, HEMOGLOBIN GAMMA CHAINS, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kidd, R.D, Baker, H.M, Mathews, A.J, Brittain, T, Baker, E.N. | | Deposit date: | 2001-02-15 | | Release date: | 2001-09-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Oligomerization and ligand binding in a homotetrameric hemoglobin: two high-resolution crystal structures of hemoglobin Bart's (gamma(4)), a marker for alpha-thalassemia.
Protein Sci., 10, 2001
|
|
