6RW2
 
 | | Bicycle Toxin Conjugate bound to EphA2 | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, ALA-ARG-ASP-CYS-PRO-LEU-VAL-ASN-PRO-LEU-CYS-LEU-HIS-PRO-GLY-TRP-THR-CYS, Ephrin type-A receptor 2, ... | | Authors: | Brown, D.G, Schroeder, S, Chen, L. | | Deposit date: | 2019-06-03 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Identification and Optimization of EphA2-Selective Bicycles for the Delivery of Cytotoxic Payloads.
J.Med.Chem., 63, 2020
|
|
5MKJ
 
 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 9 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[4-oxidanyl-3-(5-prop-2-enyl-2-propoxy-phenyl)phenyl]prop-2-enoic acid, LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-12-05 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
6RW7
 
 | | An AA10 LPMO from the shipworm symbiont Teredinibacter turnerae | | Descriptor: | COPPER (I) ION, COPPER (II) ION, Carbohydrate binding module family 33 and 10 domain protein, ... | | Authors: | Fowler, C.A, Sabbadin, F, Ciano, L, Hemsworth, G.R, Elias, L, Bruce, N.C, McQueen-Mason, S, Walton, P, Davies, G.J. | | Deposit date: | 2019-06-04 | | Release date: | 2019-10-09 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery, activity and characterisation of an AA10 lytic polysaccharide oxygenase from the shipworm symbiontTeredinibacter turnerae.
Biotechnol Biofuels, 12, 2019
|
|
5OIO
 
 | | InhA (T2A mutant) complexed with 5-((3,5-dimethyl-1H-pyrazol-1-yl)methyl)-N-ethyl-1,3,4-thiadiazol-2-amine | | Descriptor: | 5-[(3,5-dimethylpyrazol-1-yl)methyl]-~{N}-ethyl-1,3,4-thiadiazol-2-amine, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Convery, M.A. | | Deposit date: | 2017-07-19 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Screening of a Novel Fragment Library with Functional Complexity against Mycobacterium tuberculosis InhA.
ChemMedChem, 13, 2018
|
|
5OIT
 
 | |
5OJ6
 
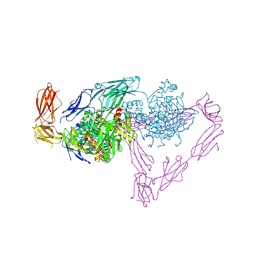 | | Crystal structure of the chicken MDGA1 ectodomain in complex with the human neuroligin 1 (NL1(-A-B)) cholinesterase domain. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAM domain-containing glycosylphosphatidylinositol anchor protein 1, Neuroligin-1, ... | | Authors: | Elegheert, J, Clayton, A.J, Aricescu, A.R. | | Deposit date: | 2017-07-20 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Mechanism for Modulation of Synaptic Neuroligin-Neurexin Signaling by MDGA Proteins.
Neuron, 95, 2017
|
|
5OKO
 
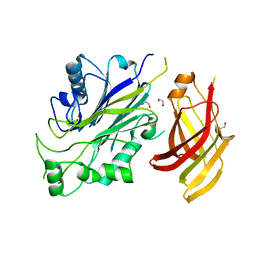 | | Crystal structure of human SHIP2 Phosphatase-C2 double mutant F593D/L597D | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 | | Authors: | Le Coq, J, Lietha, D. | | Deposit date: | 2017-07-25 | | Release date: | 2017-08-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural basis for interdomain communication in SHIP2 providing high phosphatase activity.
Elife, 6, 2017
|
|
6S0C
 
 | | Crystal structure of methionine gamma-lyase from Citrobacter freundii modified by dimethylthiosulfinate | | Descriptor: | Cystathionine gamma-synthase, PYRIDOXAL-5'-PHOSPHATE, TRIETHYLENE GLYCOL | | Authors: | Revtovich, S.V, Morozova, E.A, Nikulin, A.D, Demidkina, T.V. | | Deposit date: | 2019-06-14 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Sulfoxides of sulfur-containing amino acids are suicide substrates of Citrobacter freundii methionine gamma-lyase. Structural bases of the enzyme inactivation.
Biochimie, 168, 2020
|
|
5OKD
 
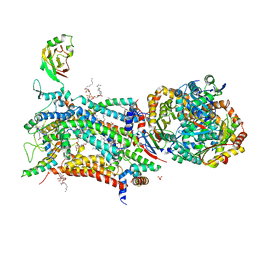 | | Crystal structure of bovine Cytochrome bc1 in complex with inhibitor SCR0911. | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Amporndanai, K, O'Neill, P.M, Hasnain, S.S, Antonyuk, S.V. | | Deposit date: | 2017-07-25 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
6S0I
 
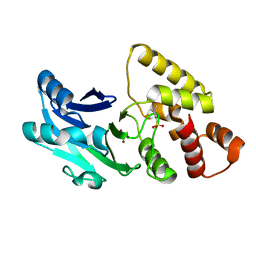 | |
5OLV
 
 | | Structure of the A2A-StaR2-bRIL562-LUAA47070 complex at 2.0A obtained from in meso soaking experiments. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-(3,3-dimethylbutanoylamino)-3,5-bis(fluoranyl)-~{N}-(1,3-thiazol-2-yl)benzamide, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Rucktooa, P, Cheng, R.K.Y, Segala, E, Geng, T, Errey, J.C, Brown, G.A, Cooke, R, Marshall, F.H, Dore, A.S. | | Deposit date: | 2017-07-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Towards high throughput GPCR crystallography: In Meso soaking of Adenosine A2A Receptor crystals.
Sci Rep, 8, 2018
|
|
5OLT
 
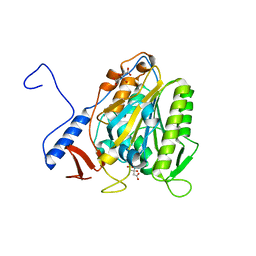 | | Crystal structure of the extramembrane domain of the cellulose biosynthetic protein BcsG from Salmonella typhimurium | | Descriptor: | CITRIC ACID, Cellulose biosynthesis protein BcsG, ZINC ION | | Authors: | Vella, P, Polyakova, A, Lindqvist, Y, Schnell, R, Bourenkov, G, Schneider, T, Schneider, G. | | Deposit date: | 2017-07-28 | | Release date: | 2018-08-08 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Characterization of the BcsG Subunit of the Cellulose Synthase in Salmonella typhimurium.
J. Mol. Biol., 430, 2018
|
|
5OCX
 
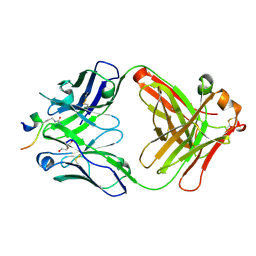 | | Crystal structure of ACPA E4 in complex with CII-C-13-CIT | | Descriptor: | 1,2-ETHANEDIOL, CII-C-13-CIT, Fab fragment anti-citrullinated protein antibody E4 - light chain, ... | | Authors: | Dobritzsch, D, Ge, C, Holmdahl, R. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Cross-Reactivity of Anti-Citrullinated Protein Antibodies.
Arthritis Rheumatol, 71, 2019
|
|
6S30
 
 | | Crystal Structure of Seryl-tRNA Synthetase from Klebsiella pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Serine--tRNA ligase | | Authors: | Pang, L, De Graef, S, Weeks, S.D, Strelkov, S.V. | | Deposit date: | 2019-06-23 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structural Insights into the Binding of Natural Pyrimidine-Based Inhibitors of Class II Aminoacyl-tRNA Synthetases.
Acs Chem.Biol., 15, 2020
|
|
5OBA
 
 | |
6S46
 
 | | Room temperature structure of the LOV2 domain of phototropin-2 from Arabidopsis thaliana 4158 ms after initiation of illumination, determined with a serial crystallography approach | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Aumonier, S, Santoni, G, Gotthard, G, von Stetten, D, Leonard, G, Royant, A. | | Deposit date: | 2019-06-26 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Millisecond time-resolved serial oscillation crystallography of a blue-light photoreceptor at a synchrotron.
Iucrj, 7, 2020
|
|
5OD3
 
 | | Crystal structure of R. ruber ADH-A, mutant Y54G, L119Y | | Descriptor: | Alcohol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Dobritzsch, D, Maurer, D, Hamnevik, E, Enugala, T.R, Widersten, M. | | Deposit date: | 2017-07-04 | | Release date: | 2018-07-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Stereo- and Regioselectivity in Catalyzed Transformation of a 1,2-Disubstituted Vicinal Diol and the Corresponding Diketone by Wild Type and Laboratory Evolved Alcohol Dehydrogenases
Acs Catalysis, 8, 2018
|
|
5ODZ
 
 | | CRYSTAL STRUCTURE OF THE BETA-LACTAMASE OXA-163 | | Descriptor: | Beta-lactamase, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Lund, B.A, Carlsen, T.J.O, Leiros, H.K.S. | | Deposit date: | 2017-07-07 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure, activity and thermostability investigations of OXA-163, OXA-181 and OXA-245 using biochemical analysis, crystal structures and differential scanning calorimetry analysis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5OE4
 
 | |
6S5Y
 
 | |
5OI9
 
 | | Trichoplax adhaerens STIL N-terminal domain | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Putative uncharacterized protein | | Authors: | van Breugel, M. | | Deposit date: | 2017-07-18 | | Release date: | 2018-04-18 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Direct binding of CEP85 to STIL ensures robust PLK4 activation and efficient centriole assembly.
Nat Commun, 9, 2018
|
|
5OIR
 
 | |
6S74
 
 | | Crystal structure of CARM1 in complex with inhibitor UM305 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-[[3-azanylpropyl-[3-(pyrimidin-2-ylamino)propyl]amino]methyl]oxolane-3,4-diol, GLYCEROL, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
6S7E
 
 | | Plant Cysteine Oxidase PCO4 from Arabidopsis thaliana (using PEG 3350 and NaF as precipitants) | | Descriptor: | FE (III) ION, Plant cysteine oxidase 4 | | Authors: | White, M.D, Levy, C.W, Flashman, E, McDonough, M.A. | | Deposit date: | 2019-07-04 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structures of Arabidopsis thaliana oxygen-sensing plant cysteine oxidases 4 and 5 enable targeted manipulation of their activity.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5OJ2
 
 | | Crystal structure of the chicken MDGA1 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAM domain-containing glycosylphosphatidylinositol anchor protein 1, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Elegheert, J, Clayton, A.J, Aricescu, A.R. | | Deposit date: | 2017-07-20 | | Release date: | 2017-08-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Mechanism for Modulation of Synaptic Neuroligin-Neurexin Signaling by MDGA Proteins.
Neuron, 95, 2017
|
|
