5IV5
 
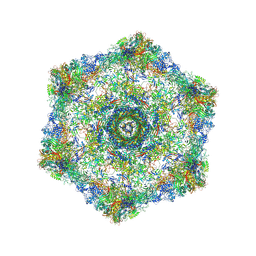 | | Cryo-electron microscopy structure of the hexagonal pre-attachment T4 baseplate-tail tube complex | | Descriptor: | Baseplate hub protein gp27, Baseplate tail-tube protein gp48, Baseplate tail-tube protein gp54, ... | | Authors: | Taylor, N.M.I, Guerrero-Ferreira, R.C, Goldie, K.N, Stahlberg, H, Leiman, P.G. | | Deposit date: | 2016-03-19 | | Release date: | 2016-05-18 | | Last modified: | 2018-02-07 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Structure of the T4 baseplate and its function in triggering sheath contraction.
Nature, 533, 2016
|
|
5IY9
 
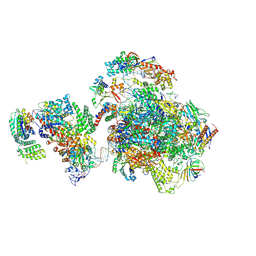 | | Human holo-PIC in the initial transcribing state (no IIS) | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
5J5H
 
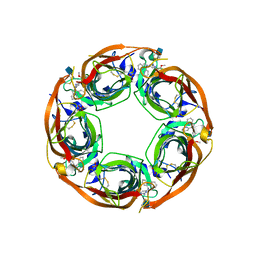 | | X-RAY STRUCTURE OF ACETYLCHOLINE BINDING PROTEIN (ACHBP) IN COMPLEX WITH 6-(2-methoxyphenyl)-N4,N4-bis[(pyridin-2-yl)methyl]pyrimidine-2,4-diamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(2-methoxyphenyl)-N~4~,N~4~-bis[(pyridin-2-yl)methyl]pyrimidine-2,4-diamine, Acetylcholine-binding protein, ... | | Authors: | Kaczanowska, K, Harel, M, Camacho Hernandez, A.G, Taylor, P. | | Deposit date: | 2016-04-02 | | Release date: | 2017-03-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substituted 2-Aminopyrimidines Selective for alpha 7-Nicotinic Acetylcholine Receptor Activation and Association with Acetylcholine Binding Proteins.
J. Am. Chem. Soc., 139, 2017
|
|
5OLY
 
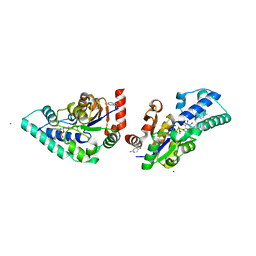 | |
6EMI
 
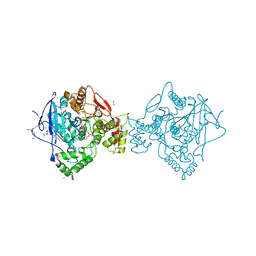 | | Crystal structure of a variant of human butyrylcholinesterase expressed in bacteria. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cholinesterase, ... | | Authors: | Brazzolotto, X, Igert, A, Guillon, V, Santoni, G, Nachon, F. | | Deposit date: | 2017-10-02 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.476 Å) | | Cite: | Bacterial Expression of Human Butyrylcholinesterase as a Tool for Nerve Agent Bioscavengers Development.
Molecules, 22, 2017
|
|
6EMW
 
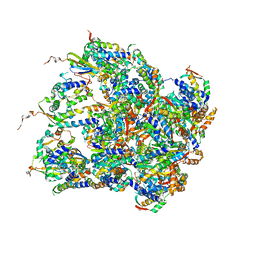 | | Structure of S.aureus ClpC in complex with MecA | | Descriptor: | ATP-dependent Clp protease ATP-binding subunit, ATP-dependent Clp protease ATP-binding subunit ClpC, Adapter protein MecA, ... | | Authors: | Carroni, M, Mogk, A, Bukau, B, Franke, K. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Regulatory coiled-coil domains promote head-to-head assemblies of AAA+ chaperones essential for tunable activity control.
Elife, 6, 2017
|
|
6P5N
 
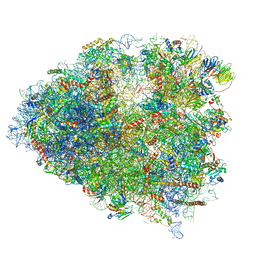 | | Structure of a mammalian 80S ribosome in complex with a single translocated Israeli Acute Paralysis Virus IRES and eRF1 | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-25 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
5OPQ
 
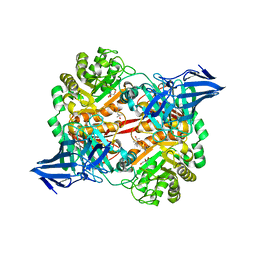 | | A 3,6-anhydro-D-galactosidase produced by Zobellia galactanivorans. This is an exo-lytic enzyme that hydrolyzes terminal 3,6-anhydro-D-galactose from the non-reducing end of carrageenan oligosaccharides. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6-anhydro-D-galactosidase, ... | | Authors: | Ficko-Blean, E, Michel, G, Czjzek, M. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Carrageenan catabolism is encoded by a complex regulon in marine heterotrophic bacteria.
Nat Commun, 8, 2017
|
|
6EP2
 
 | |
5OP3
 
 | | Designed Ankyrin Repeat Protein (DARPin) NDNH-1 selected by directed evolution against Lysozyme | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fischer, G, Hogan, B.J, Houlihan, G, Edmond, S, Huovinen, T.T.K, Hollfelder, F, Hyvonen, M. | | Deposit date: | 2017-08-09 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.359 Å) | | Cite: | Designed Ankyrin Repeat Protein (DARPin) NDNH-1 selected by directed evolution against Lysozyme
To be published
|
|
6E2X
 
 | | Mechanism of cellular recognition by PCV2 | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Capsid protein of PCV2 | | Authors: | Khayat, R, Dhindwal, S. | | Deposit date: | 2018-07-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Porcine Circovirus 2 Uses a Multitude of Weak Binding Sites To Interact with Heparan Sulfate, and the Interactions Do Not Follow the Symmetry of the Capsid.
J.Virol., 93, 2019
|
|
6E6B
 
 | | Crystal structure of the Protocadherin GammaB4 extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2018-07-24 | | Release date: | 2019-04-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.52 Å) | | Cite: | Visualization of clustered protocadherin neuronal self-recognition complexes.
Nature, 569, 2019
|
|
5OQF
 
 | |
8U8Q
 
 | | V290N/S292F Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6E9D
 
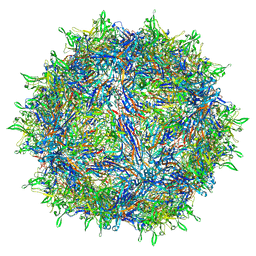 | | Sub-2 Angstrom Ewald Curvature-Corrected Single-Particle Cryo-EM Reconstruction of AAV-2 L336C | | Descriptor: | Capsid protein VP1 | | Authors: | Tan, Y.Z, Aiyer, S, Mietzsch, M, Hull, J.A, McKenna, R, Baker, T.S, Agbandje-McKenna, M, Lyumkis, D. | | Deposit date: | 2018-07-31 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (1.86 Å) | | Cite: | Sub-2 angstrom Ewald curvature corrected structure of an AAV2 capsid variant.
Nat Commun, 9, 2018
|
|
5JUU
 
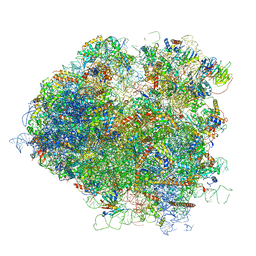 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure V (least rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2019-11-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
8U8P
 
 | | S292F Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8U8R
 
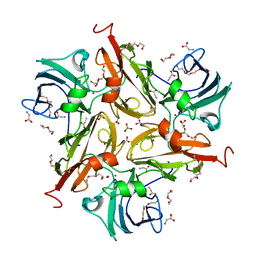 | | Y229F/V290N/S292F Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8U28
 
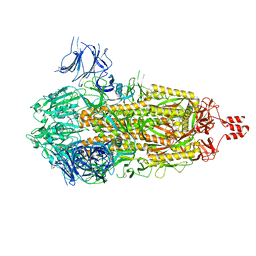 | |
8U8S
 
 | | Y229F/S292F Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8U8T
 
 | | Y229F/V290N Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6E2Z
 
 | | Mechanism of cellular recognition by PCV2 | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Capsid protein of PCV2 | | Authors: | Khayat, R, Dhindwal, S. | | Deposit date: | 2018-07-12 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Porcine Circovirus 2 Uses a Multitude of Weak Binding Sites To Interact with Heparan Sulfate, and the Interactions Do Not Follow the Symmetry of the Capsid.
J.Virol., 93, 2019
|
|
8U10
 
 | | In situ cryo-EM structure of bacteriophage P22 gp1:gp4:gp5:gp10:gp9 N-term complex in conformation 1 at 3.2A resolution | | Descriptor: | Major capsid protein, Packaged DNA stabilization protein gp10, Peptidoglycan hydrolase gp4, ... | | Authors: | Iglesias, S, Feng-Hou, C, Cingolani, G. | | Deposit date: | 2023-08-30 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular Architecture of Salmonella Typhimurium Virus P22 Genome Ejection Machinery.
J.Mol.Biol., 435, 2023
|
|
6PJW
 
 | | Adenylate kinase from Methanococcus igneus - AMP bound form | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenylate kinase, MAGNESIUM ION | | Authors: | Moon, S, Kim, J, Olmos, J.L, Bae, E, Phillips Jr, G.N. | | Deposit date: | 2019-06-28 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Adenylate kinase from Methanococcus igneus - AMP bound form
To Be Published
|
|
8UH4
 
 | |
