6SYO
 
 | | Hydrogenase-2 variant R479K - As Isolated form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2019-09-30 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Hydrogen activation by NiFe-hydrogenases - consolidating the role of the pendant arginine.
To Be Published
|
|
5OFY
 
 | | Crystal structure of the D183N variant of human Alanine:Glyoxylate Aminotransferase major allele (AGT-Ma) at pH 9.0. 2.8 Ang; internal aldimine with PLP in the active site | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, PYRIDOXAL-5'-PHOSPHATE, Serine--pyruvate aminotransferase | | Authors: | Giardina, G, Cutruzzola, F, Borri Voltattorni, C, Cellini, B, Montioli, R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Radiation damage at the active site of human alanine:glyoxylate aminotransferase reveals that the cofactor position is finely tuned during catalysis.
Sci Rep, 7, 2017
|
|
6SZK
 
 | | Hydrogenase-2 variant R479K - hydrogen reduced form treated with CO | | Descriptor: | CARBON MONOXIDE, CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | Authors: | Carr, S.B, Beaton, S.E, Evans, R.M, Armstrong, F.A. | | Deposit date: | 2019-10-02 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hydrogen activation by NiFe-hydrogenases - consolidating the role of the pendant arginine.
To Be Published
|
|
5OG4
 
 | | Cu nitrite reductase serial data at varying temperatures RT 0.18MGy | | Descriptor: | ACETATE ION, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Horrell, S, Kekilli, D, Strange, R.W, Hough, M.A. | | Deposit date: | 2017-07-11 | | Release date: | 2018-05-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Enzyme catalysis captured using multiple structures from one crystal at varying temperatures.
IUCrJ, 5, 2018
|
|
6S9E
 
 | | Tubulin-GDP.AlF complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALUMINUM FLUORIDE, CALCIUM ION, ... | | Authors: | Oliva, M.A, Estevez-Gallego, J, Diaz, J.F, Prota, A.E, Steinmetz, M.O, Balaguer, F.A, Lucena-Agell, D. | | Deposit date: | 2019-07-12 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural model for differential cap maturation at growing microtubule ends.
Elife, 9, 2020
|
|
5NVW
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(cyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 6) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-(cyclopropylcarbonylamino)-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
6SE3
 
 | | Crystal Structure of Ancestral Flavin-containing monooxygenase (FMO) 3-6 | | Descriptor: | Ancestral Flavin-containing monooxygenase (FMO) 3-6, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Nicoll, C, Bailleul, G, Fiorentini, F, Mascotti, M.L, Fraaije, M, Mattevi, A. | | Deposit date: | 2019-07-29 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ancestral-sequence reconstruction unveils the structural basis of function in mammalian FMOs.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5NSG
 
 | | Fc DEDE homodimer variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Putative uncharacterized protein DKFZp686C11235, ... | | Authors: | De Nardis, C, Hendriks, L.J.A, Poirier, E, Arvinte, T, Gros, P, Bakker, A.B.H, de Kruif, J. | | Deposit date: | 2017-04-26 | | Release date: | 2017-07-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A new approach for generating bispecific antibodies based on a common light chain format and the stable architecture of human immunoglobulin G1.
J. Biol. Chem., 292, 2017
|
|
6SFO
 
 | | MAPK14 with bound inhibitor SR-318 | | Descriptor: | 5-azanyl-~{N}-[[4-(3-cyclohexylpropylcarbamoyl)phenyl]methyl]-1-phenyl-pyrazole-4-carboxamide, GLYCEROL, Mitogen-activated protein kinase 14, ... | | Authors: | Schroeder, M, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-08-01 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | MAPK14 with bound inhibitor SR-318
To Be Published
|
|
5NSK
 
 | | apo Structure of Leucyl aminopeptidase from Trypanosoma brucei | | Descriptor: | Aminopeptidase, putative | | Authors: | Timm, J, Wilson, K.S. | | Deposit date: | 2017-04-26 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Characterization of Acidic M17 Leucine Aminopeptidases from the TriTryps and Evaluation of Their Role in Nutrient Starvation inTrypanosoma brucei.
mSphere, 2, 2018
|
|
5NTF
 
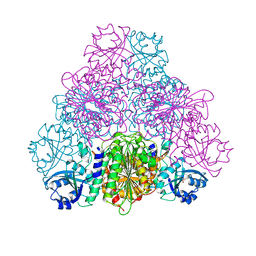 | | apo Structure of Leucyl aminopeptidase from Trypanosoma cruzi | | Descriptor: | Aminopeptidase, SULFATE ION | | Authors: | Timm, J, Wilson, K. | | Deposit date: | 2017-04-27 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Characterization of Acidic M17 Leucine Aminopeptidases from the TriTryps and Evaluation of Their Role in Nutrient Starvation inTrypanosoma brucei.
mSphere, 2, 2017
|
|
6SF4
 
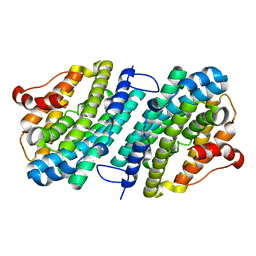 | | Apo form of the ribonucleotide reductase NrdB protein from Leeuwenhoekiella blandensis | | Descriptor: | Ribonucleoside-diphosphate reductase, beta subunit 1 | | Authors: | Hasan, M, Rozman Grinberg, I, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2019-07-31 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Class Id ribonucleotide reductase utilizes a Mn2(IV,III) cofactor and undergoes large conformational changes on metal loading.
J.Biol.Inorg.Chem., 24, 2019
|
|
5NTT
 
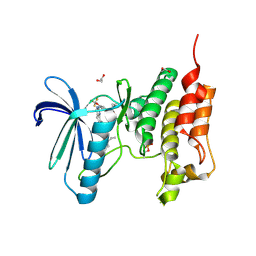 | | Crystal structure of human Mps1 (TTK) C604Y mutant in complex with NMS-P715 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity protein kinase TTK, N-(2,6-DIETHYLPHENYL)-1-METHYL-8-({4-[(1-METHYLPIPERIDIN-4-YL)CARBAMOYL]-2-(TRIFLUOROMETHOXY)PHENYL}AMINO)-4,5-DIHYDRO-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE | | Authors: | Hiruma, Y, Joosten, R.P, Perrakis, A. | | Deposit date: | 2017-04-28 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Understanding inhibitor resistance in Mps1 kinase through novel biophysical assays and structures.
J. Biol. Chem., 292, 2017
|
|
6SFH
 
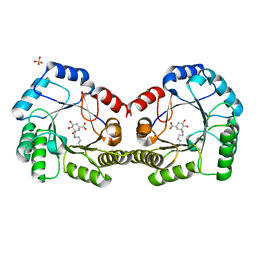 | | CRYSTAL STRUCTURE OF DHQ1 FROM Staphylococcus aureus COVALENTLY MODIFIED BY LIGAND 7 | | Descriptor: | (1~{S},3~{S},4~{S},5~{R})-3-(aminomethyl)-3,4,5-tris(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase, LITHIUM ION, ... | | Authors: | Sanz-Gaitero, M, Lence, E, Maneiro, M, Thompson, R, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2019-08-01 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Self-Immolation of a Bacterial Dehydratase Enzyme by its Epoxide Product.
Chemistry, 26, 2020
|
|
5NU5
 
 | |
5NUE
 
 | | Cytosolic Malate Dehydrogenase 1 (peroxide-treated) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-ETHOXYETHANOL, ... | | Authors: | Young, D, Messens, J, Huang, J, Reichheld, J.-P. | | Deposit date: | 2017-04-29 | | Release date: | 2018-02-28 | | Last modified: | 2018-07-04 | | Method: | X-RAY DIFFRACTION (1.35000277 Å) | | Cite: | Self-protection of cytosolic malate dehydrogenase against oxidative stress in Arabidopsis.
J. Exp. Bot., 69, 2018
|
|
5NVD
 
 | | Crystal structure of hexameric CBS-CP12 protein from bloom-forming cyanobacteria at 2.5 A resolution in P6322 crystal form | | Descriptor: | CBS-CP12 | | Authors: | Hackenberg, C, Hakanpaa, J, Eigner, C, Antonyuk, S.V, Dittmann, E, Lamzin, V.S. | | Deposit date: | 2017-05-04 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional insights into the unique CBS-CP12 fusion protein family in cyanobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5NW5
 
 | | Crystal structure of the Rif1 N-terminal domain (RIF1-NTD) from Saccharomyces cerevisiae in complex with DNA | | Descriptor: | DNA (30-MER), DNA (60-MER), Telomere length regulator protein RIF1 | | Authors: | Bunker, R.D, Reinert, J.K, Shi, T, Thoma, N.H. | | Deposit date: | 2017-05-05 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (6.502 Å) | | Cite: | Rif1 maintains telomeres and mediates DNA repair by encasing DNA ends.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5NXL
 
 | | Formylglycine generating enzyme from T. curvata in complex with Ag(I) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Meury, M, Knop, M, Seebeck, F.P. | | Deposit date: | 2017-05-10 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural Basis for Copper-Oxygen Mediated C-H Bond Activation by the Formylglycine-Generating Enzyme.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
6SMT
 
 | | S-enantioselective imine reductase from Mycobacterium smegmatis | | Descriptor: | (2S)-2-ethylhexan-1-ol, 1,2-ETHANEDIOL, 6-phosphogluconate dehydrogenase, ... | | Authors: | Meyer, T, Zumbraegel, N, Geerds, C, Groeger, H, Niemann, H.H. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Characterization of an S -enantioselective Imine Reductase from Mycobacterium Smegmatis .
Biomolecules, 10, 2020
|
|
6SNE
 
 | | crystal structure of LN01 Fab in complex with an HIV-1 gp41 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, LN01 heavy chain, ... | | Authors: | Caillat, C, Pinto, D, Corti, D, Fenwick, C, Pantaleo, G, Weissenhorn, W. | | Deposit date: | 2019-08-23 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural Basis for Broad HIV-1 Neutralization by the MPER-Specific Human Broadly Neutralizing Antibody LN01.
Cell Host Microbe, 26, 2019
|
|
6SKR
 
 | | OXA-10_ETP. Structural insight to the enhanced carbapenem efficiency of OXA-655 compared to OXA-10. | | Descriptor: | (4R,5S)-3-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, FORMIC ACID, ... | | Authors: | Leiros, H.-K.S. | | Deposit date: | 2019-08-16 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural insights into the enhanced carbapenemase efficiency of OXA-655 compared to OXA-10.
Febs Open Bio, 10, 2020
|
|
5O1L
 
 | | Structure of Latex Clearing Protein LCP in the open state with bound imidazole | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 1,2-ETHANEDIOL, IMIDAZOLE, ... | | Authors: | Ilcu, L, Roether, W, Birke, J, Brausemann, A, Einsle, O, Jendrossek, D. | | Deposit date: | 2017-05-18 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and Functional Analysis of Latex Clearing Protein (Lcp) Provides Insight into the Enzymatic Cleavage of Rubber.
Sci Rep, 7, 2017
|
|
6SPK
 
 | | A4V MUTANT OF HUMAN SOD1 WITH EBSELEN DERIVATIVE 6 | | Descriptor: | 2-selanyl-~{N}-[3-[4-(trifluoromethyl)phenyl]phenyl]benzamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Chantadul, V, Amporndanai, K, Wright, G, Shahid, M, Antonyuk, S, Hasnain, S. | | Deposit date: | 2019-09-01 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Ebselen as template for stabilization of A4V mutant dimer for motor neuron disease therapy.
Commun Biol, 3, 2020
|
|
5NSM
 
 | | unliganded Structure of Leucyl aminopeptidase from Trypanosoma brucei | | Descriptor: | Aminopeptidase, putative, BICARBONATE ION, ... | | Authors: | Timm, J, Wilson, K. | | Deposit date: | 2017-04-26 | | Release date: | 2017-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Characterization of Acidic M17 Leucine Aminopeptidases from the TriTryps and Evaluation of Their Role in Nutrient Starvation inTrypanosoma brucei.
mSphere, 2, 2018
|
|
