6UAI
 
 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM complexed with YSAM peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
7AMG
 
 | | IL-17A in complex with small molecule modulators | | Descriptor: | (3~{R})-4-[4-[[(2~{S})-2-[[2,2-bis(fluoranyl)-2-phenyl-ethanoyl]amino]-3-(2-chlorophenyl)propanoyl]amino]phenyl]-3-[[(2~{S})-3-methyl-2-[2-[2-[(2-methylpropan-2-yl)oxycarbonylamino]ethoxy]ethanoylamino]butanoyl]amino]butanoic acid, Interleukin-17A | | Authors: | Hakansson, M, Kimbung, R, Logan, D, Walse, U.B, de Groot, M.J, Andrews, M.D, Dack, K.N, Lambert, M. | | Deposit date: | 2020-10-08 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Discovery of an Oral, Rule of 5 Compliant, Interleukin 17A Protein-Protein Interaction Modulator for the Potential Treatment of Psoriasis and Other Inflammatory Diseases.
J.Med.Chem., 65, 2022
|
|
5NVZ
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(1-acetylcyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 16) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-ethanoylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
6U72
 
 | | BRD4-BD1 in complex with the cyclic peptide 3.1_2_AcK5toA | | Descriptor: | 3.1_2_AcK5toA, AMINO GROUP, Bromodomain-containing protein 4 | | Authors: | Patel, K, Walshe, J.L, Walport, L.J, Mackay, J.P, Mouradian, K.S. | | Deposit date: | 2019-08-31 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cyclic peptides can engage a single binding pocket through highly divergent modes.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5NUR
 
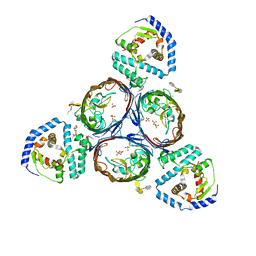 | | Structural basis for maintenance of bacterial outer membrane lipid asymmetry | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-beta-D-glucopyranose-(1-6)-2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(3-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-beta-D-glucopyranose-(1-6)-2-amino-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, ... | | Authors: | Abellon-Ruiz, J, Kaptan, S.S, Basle, A, Claudi, B, Bumann, D, Kleinekathofer, U, van den Berg, B. | | Deposit date: | 2017-05-01 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural basis for maintenance of bacterial outer membrane lipid asymmetry.
Nat Microbiol, 2, 2017
|
|
7AMU
 
 | | Crystal structure of rsEGFP2 T204A in its fluorescent on-state | | Descriptor: | GLYCEROL, Green fluorescent protein, SULFATE ION | | Authors: | Moreno-Chicano, T, Schlichting, I, Hartmann, E, Zala, N, Colletier, J.-P, Weik, M. | | Deposit date: | 2020-10-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structure of rsEGFP2 in its fluorescent on-state at pH 8.0
To Be Published
|
|
5NVG
 
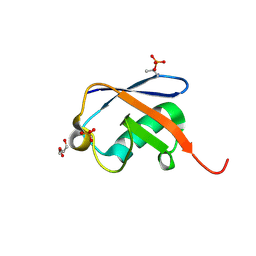 | | Thr12 Phosphorylated Ubiquitin | | Descriptor: | DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, Polyubiquitin-B | | Authors: | Huguenin-Dezot, N. | | Deposit date: | 2017-05-04 | | Release date: | 2017-05-31 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Biosynthesis and genetic encoding of phosphothreonine through parallel selection and deep sequencing.
Nat. Methods, 14, 2017
|
|
6U8C
 
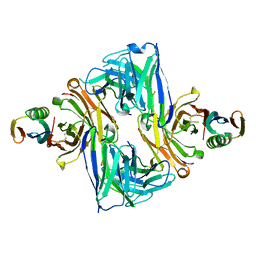 | | Crystal structure of an engineered ultra-high affinity Fab-Protein G complex | | Descriptor: | Antibody heavy chain Fab, Antibody light chain Fab, Protein G | | Authors: | Slezak, T, Filippova, E.V, Davydova, E.K, Kossiakoff, A.A. | | Deposit date: | 2019-09-04 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | An engineered ultra-high affinity Fab-Protein G pair enables a modular antibody platform with multifunctional capability.
Protein Sci., 29, 2020
|
|
7AYY
 
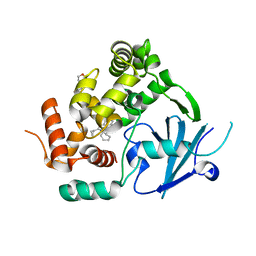 | | Structure of the human 8-oxoguanine DNA Glycosylase hOGG1 in complex with activator TH10785 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Davies, J.R, Stenmark, P. | | Deposit date: | 2020-11-13 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-molecule activation of OGG1 increases oxidative DNA damage repair by gaining a new function.
Science, 376, 2022
|
|
5NYK
 
 | | Crystal structure of the atypical poplar thioredoxin-like2.1 in oxidized state | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Chibani, K, Saul, F.A, Haouz, A, Rouhier, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural snapshots along the reaction mechanism of the atypical poplar thioredoxin-like2.1.
FEBS Lett., 592, 2018
|
|
5NW0
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(1-acetamidocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 17) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-[(1-acetamidocyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
7AYC
 
 | | Crystal Structure of human mitochondrial 2-Enoyl Thioester Reductase (MECR) with single mutation G165Q | | Descriptor: | CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase, mitochondrial | | Authors: | Rahman, M.T, Koski, M.K, Autio, K.J, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2020-11-12 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | An engineered variant of MECR reductase reveals indispensability of long-chain acyl-ACPs for mitochondrial respiration.
Nat Commun, 14, 2023
|
|
7AVN
 
 | | Structure of marine actinobacteria clade rhodopsin (MacR) in orange form in P1 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, ... | | Authors: | Gushchin, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gordeliy, V. | | Deposit date: | 2020-11-05 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Alternating Access Mechanism in a Minimalistic Light-Driven Proton Pump
To Be Published
|
|
7AYZ
 
 | |
5NZY
 
 | | Crystal structure of DNA cross-link repair protein 1A in complex with Cefotaxime | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, DNA cross-link repair 1A protein, NICKEL (II) ION | | Authors: | Newman, J.A, Aitkenhead, H, Kupinska, K, Burgess-Brown, N.A, Talon, R, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2017-05-15 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.551 Å) | | Cite: | Crystal structure of DNA cross-link repair protein 1A in complex with Cefotaxime
To Be Published
|
|
7AZ0
 
 | |
7AYB
 
 | | Crystal Structure of wild type human mitochondrial 2-Enoyl Thioester Reductase (MECR) | | Descriptor: | ACETIC ACID, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase, ... | | Authors: | Rahman, M.T, Koski, M.K, Autio, K.J, Kastaniotis, A.J, Wierenga, R.K, Hiltunen, J.K. | | Deposit date: | 2020-11-12 | | Release date: | 2022-06-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An engineered variant of MECR reductase reveals indispensability of long-chain acyl-ACPs for mitochondrial respiration.
Nat Commun, 14, 2023
|
|
7AVO
 
 | | Structure of marine actinobacteria clade rhodopsin (MacR) in orange form in P1211 space group | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Bacteriorhodopsin, ... | | Authors: | Gushchin, I, Polovinkin, V, Kovalev, K, Shevchenko, V, Gordeliy, V. | | Deposit date: | 2020-11-05 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Alternating Access Mechanism in a Minimalistic Light-Driven Proton Pump
To Be Published
|
|
6UAO
 
 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM complexed with the peptide EEYSAM | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptide EEYSAM, ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
7B1T
 
 | | Crystal structure of BRD4(1) in complex with the inhibitor MPM6 | | Descriptor: | 3-(5-azanyl-2-chloranyl-phenyl)-1-methyl-4,7-dihydro-2~{H}-cyclohepta[c]pyrrol-8-one, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE | | Authors: | Huegle, M. | | Deposit date: | 2020-11-25 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A novel pan-selective bromodomain inhibitor for epigenetic drug design.
Eur.J.Med.Chem., 249, 2023
|
|
6TV9
 
 | |
5NX0
 
 | | Structure of Spin-labelled T4 lysozyme mutant L115C-R119C-R1 at room temperature | | Descriptor: | Endolysin | | Authors: | Gohlke, U, Loll, B, Consentius, P, Mueller, R, Kaupp, M, Heinemann, U, Wahl, M.C, Risse, T. | | Deposit date: | 2017-05-09 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Combining EPR spectroscopy and X-ray crystallography to elucidate the structure and dynamics of conformationally constrained spin labels in T4 lysozyme single crystals.
Phys Chem Chem Phys, 19, 2017
|
|
7B55
 
 | | Crystal structure of CaMKII-actinin complex bound to MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-actinin-2, Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | Zhu, J, Gold, M. | | Deposit date: | 2020-12-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of CaMKII-actinin complex bound to MES
To Be Published
|
|
6TWP
 
 | | Binding domain of BoNT/A5 | | Descriptor: | Botulinum neurotoxin A5, CHLORIDE ION | | Authors: | Davies, J.R, Acharya, K.R. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High-resolution crystal structures of the botulinum neurotoxin binding domains from subtypes A5 and A6.
Febs Open Bio, 10, 2020
|
|
7B56
 
 | | Crystal structure of CaMKII-actinin complex bound to AMPPNP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-actinin-2, Calcium/calmodulin-dependent protein kinase type II subunit alpha, ... | | Authors: | Zhu, J, Gold, M. | | Deposit date: | 2020-12-03 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of CaMKII-actinin complex bound to MES
To Be Published
|
|
