5TT4
 
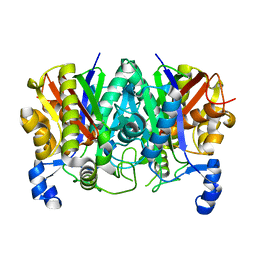 | | Determining the Molecular Basis For Starter Unit Selection During Daunorubicin Biosynthesis | | Descriptor: | Daunorubicin-doxorubicin polyketide synthase | | Authors: | Jackson, D.R, Valentic, T.R, Patel, A, Tsai, S.C, Mohammed, L, Vasilakis, K, Wattana-amorn, P, Long, P.F, Crump, M.P, Crosby, J. | | Deposit date: | 2016-11-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determining the Molecular Basis For Starter Unit Selection During Daunorubicin Biosynthesis
To Be Published
|
|
5TVL
 
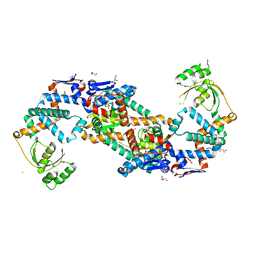 | | Crystal structure of foldase protein PrsA from Streptococcus pneumoniae str. Canada MDR_19A | | Descriptor: | CHLORIDE ION, Foldase protein PrsA, GLYCEROL, ... | | Authors: | Borek, D, Yim, V, Kudritska, M, Wawrzak, Z, Stogios, P.J, Otwinowski, Z, Savchenko, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-09 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of foldase protein PrsA from Streptococcus pneumoniae str. Canada MDR_19A
To Be Published
|
|
3T78
 
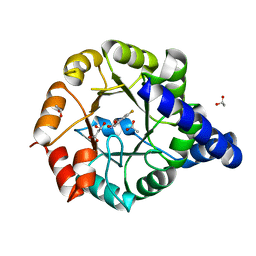 | | Crystal Structure of Mycobacterium tuberculosis Indole Glycerol Phosphate Synthase (IGPS) in Complex with 5-Fluoroanthranilate | | Descriptor: | 2-amino-5-fluorobenzoic acid, ACETATE ION, Indole-3-glycerol phosphate synthase, ... | | Authors: | Reddy, M.C.M, Bruning, J.B, Thurman, C, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-07-29 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights and Inhibition of Mycobacterium tuberculosis Indole Glycerol Phosphate Synthase (IGPS): an Essential Enzyme for Tryptophan Biosynthesis
To be Published
|
|
5UDN
 
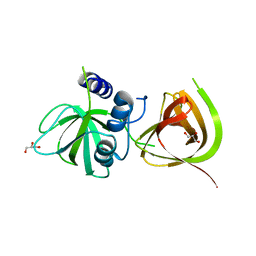 | |
3E0R
 
 | | Crystal structure of cppA protein from Streptococcus pneumoniae TIGR4 | | Descriptor: | C3-degrading proteinase (CppA protein), CHLORIDE ION | | Authors: | Nocek, B, Mulligan, R, Abdullah, J, Otwinowski, Z, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-31 | | Release date: | 2008-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of cppA protein from Streptococcus pneumoniae TIGR4
To be Published
|
|
1D2R
 
 | |
5U9Z
 
 | |
3T40
 
 | | Crystal Structure of Mycobacterium tuberculosis Indole Glycerol Phosphate Synthase (IGPS) complex with N-2-Carboxyphenyl Glycine (CPG) | | Descriptor: | 2-[(carboxymethyl)amino]benzoic acid, Indole-3-glycerol phosphate synthase, SULFATE ION | | Authors: | Reddy, M.C.M, Bruning, J.B, Thurman, C, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-07-25 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Structural Insights and Inhibition of Mycobacterium tuberculosis Indole Glycerol Phosphate Synthase (IGPS): an Essential Enzyme for Tryptophan Biosynthesis
To be Published
|
|
3TC6
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR63. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
5U8K
 
 | | RitR mutant - C128S | | Descriptor: | Response regulator | | Authors: | Silvaggi, N.R, Han, L. | | Deposit date: | 2016-12-14 | | Release date: | 2017-12-20 | | Last modified: | 2018-11-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | RitR is an archetype for a novel family of redox sensors in the streptococci that has evolved from two-component response regulators and is required for pneumococcal colonization.
PLoS Pathog., 14, 2018
|
|
5UDM
 
 | |
1DL3
 
 | | CRYSTAL STRUCTURE OF MUTUALLY GENERATED MONOMERS OF DIMERIC PHOSPHORIBOSYLANTRANILATE ISOMERASE FROM THERMOTOGA MARITIMA | | Descriptor: | PROTEIN (PHOSPHORIBOSYLANTRANILATE ISOMERASE), SULFATE ION | | Authors: | Thoma, R, Hennig, M, Sterner, R, Kirschner, K. | | Deposit date: | 1999-12-08 | | Release date: | 1999-12-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of mutationally generated monomers of dimeric phosphoribosylanthranilate isomerase from Thermotoga maritima.
Structure Fold.Des., 8, 2000
|
|
5UNC
 
 | | The crystal structure of PHOSPHOENOLPYRUVATE PHOSPHOMUTASE from Streptomyces platensis subsp. rosaceus | | Descriptor: | FORMIC ACID, L(+)-TARTARIC ACID, PHOSPHOENOLPYRUVATE PHOSPHOMUTASE, ... | | Authors: | Tan, K, Hatzos-Skintges, C, Endres, M, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-01-30 | | Release date: | 2017-02-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | The crystal structure of PHOSPHOENOLPYRUVATE PHOSPHOMUTASE from Streptomyces platensis subsp. rosaceus
To Be Published
|
|
3AIG
 
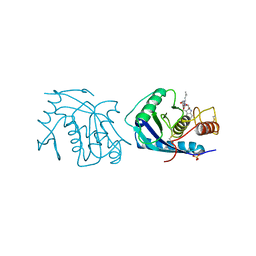 | | ADAMALYSIN II WITH PEPTIDOMIMETIC INHIBITOR POL656 | | Descriptor: | (3R)-2-[N-(furan-2-ylcarbonyl)-L-leucyl]-2,3,4,9-tetrahydro-1H-beta-carboline-3-carboxylic acid, ADAMALYSIN II, CALCIUM ION, ... | | Authors: | Gomis-Rueth, F.X, Meyer, E.F, Kress, L.F, Politi, V. | | Deposit date: | 1997-10-12 | | Release date: | 1998-04-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of adamalysin II with peptidic inhibitors. Implications for the design of tumor necrosis factor alpha convertase inhibitors.
Protein Sci., 7, 1998
|
|
5UPQ
 
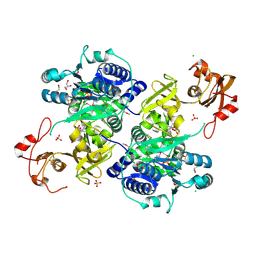 | | Acyl-CoA synthetase PtmA2 from Streptomyces platensis in complex with SBNP465 ligand | | Descriptor: | 5'-O-[(R)-{[(7beta,8alpha,9beta,10alpha,13alpha,16beta)-7,16-dihydroxy-18-oxokauran-18-yl]oxy}(hydroxy)phosphoryl]adenosine, Acyl-CoA synthetase PtmA2, CHLORIDE ION, ... | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J.D, Chang, C.Y, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-02-03 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme.
Nat. Chem. Biol., 14, 2018
|
|
5UPS
 
 | | Acyl-CoA synthetase PtmA2 from Streptomyces platensis in complex with SBNP663 ligand | | Descriptor: | 5'-O-[(R)-hydroxy{[(7beta,8alpha,9beta,10alpha,11beta,13alpha)-7-hydroxy-19-oxo-11,16-epoxykauran-19-yl]oxy}phosphoryl]adenosine, Acyl-CoA synthetase PtmA2, FORMIC ACID, ... | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J.D, Chang, C.-Y, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-02-03 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme.
Nat. Chem. Biol., 14, 2018
|
|
3EIF
 
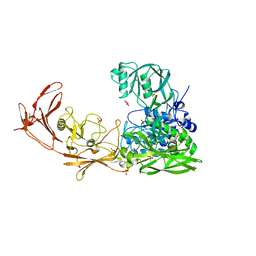 | | 1.9 angstrom crystal structure of the active form of the C5a peptidase from Streptococcus pyogenes (ScpA) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, C5a peptidase, CALCIUM ION, ... | | Authors: | Cooney, J.C, Kagawa, T.F, O'Connell, M.R, Paoli, M, Mouat, P, O'Toole, P.W. | | Deposit date: | 2008-09-15 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Model for Substrate Interactions in C5a Peptidase from Streptococcus pyogenes: A 1.9 A Crystal Structure of the Active Form of ScpA
J.Mol.Biol., 386, 2009
|
|
4HKM
 
 | |
8UUB
 
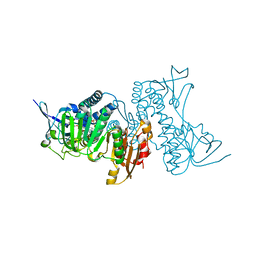 | | Structure of hypothiocyanous acid reductase (Har) from Streptococcus pneumoniae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MALONATE ION, Oxidoreductase, ... | | Authors: | Shearer, H.L, Currie, M.J, Dickerhof, N, Dobson, R.C.J. | | Deposit date: | 2023-10-31 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Hypothiocyanous acid reductase is critical for host colonization and infection by Streptococcus pneumoniae.
J.Biol.Chem., 300, 2024
|
|
4YW4
 
 | | Streptococcus pneumoniae sialidase NanC | | Descriptor: | GLYCEROL, Neuraminidase C, PHOSPHATE ION | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-03-20 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a novel sialidase
To be published
|
|
8YT5
 
 | | SP1746 treated with EDTA, in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FE (III) ION, bis(5'-nucleosyl)-tetraphosphatase (symmetrical) | | Authors: | Jin, Y, Niu, L, Ke, J. | | Deposit date: | 2024-03-25 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and biochemical characterization of a nucleotide hydrolase from Streptococcus pneumonia.
Structure, 2024
|
|
8TGT
 
 | |
4YW1
 
 | | Crystal Structure of Streptococcus pneumoniae NanC, complex with Neu5Ac and Neu5Ac2en following soaking with 3'SL | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Owen, C.D, Lukacik, P, Potter, J.A, Walsh, M, Taylor, G.L. | | Deposit date: | 2015-03-20 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Streptococcus pneumoniae NanC: STRUCTURAL INSIGHTS INTO THE SPECIFICITY AND MECHANISM OF A SIALIDASE THAT PRODUCES A SIALIDASE INHIBITOR.
J.Biol.Chem., 290, 2015
|
|
8WMY
 
 | | Crystal structure of YqeK in complex with MG and ADP. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, bis(5'-nucleosyl)-tetraphosphatase (symmetrical) | | Authors: | Mo, X. | | Deposit date: | 2023-10-05 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Molecular Mechanism of Ap4A Hydrolysis by YqeK of Staphylococcus pyogenes.
To be published
|
|
5GZJ
 
 | | Cyclodeaminase_PA | | Descriptor: | Lysine cyclodeaminase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Ying, H, Chen, K. | | Deposit date: | 2016-09-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Cyclodeaminase_PA
To Be Published
|
|
