2DQ0
 
 | | Crystal structure of seryl-tRNA synthetase from Pyrococcus horikoshii complexed with a seryl-adenylate analog | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, SULFATE ION, Seryl-tRNA synthetase | | Authors: | Itoh, Y, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-18 | | Release date: | 2007-06-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic and mutational studies of seryl-tRNA synthetase from the archaeon Pyrococcus horikoshii.
Rna Biol., 5, 2008
|
|
3BIH
 
 | | Crystal structure of fructose-1,6-bisphosphatase from E.coli GlpX | | Descriptor: | Fructose-1,6-bisphosphatase class II glpX, UNKNOWN ATOM OR ION | | Authors: | Lunin, V.V, Skarina, T, Brown, G, Yakunin, A.F, Edwards, A.M, Savchenko, A. | | Deposit date: | 2007-11-30 | | Release date: | 2008-12-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
4HBW
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazoline ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N-ethyl-3-methyl-2-oxo-1,2,3,4-tetrahydroquinazoline-6-sulfonamide, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
4GPJ
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a isoxazolylbenzimidazole ligand | | Descriptor: | (1R)-6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-phenyl-2,3-dihydro-1H-inden-1-ol, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Heightman, T.D, Brennan, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The design and synthesis of 5- and 6-isoxazolylbenzimidazoles as selective inhibitors of the BET bromodomains.
Medchemcomm, 4, 2013
|
|
4HBY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolin ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-2-oxo-N-phenyl-1,2,3,4-tetrahydroquinazoline-6-sulfonamide, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
2Z4G
 
 | | Histidinol Phosphate Phosphatase from Thermus thermophilus HB8 | | Descriptor: | FE (III) ION, GLYCEROL, Histidinol phosphatase, ... | | Authors: | Omi, R. | | Deposit date: | 2007-06-17 | | Release date: | 2007-11-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Monofunctional Histidinol Phosphate Phosphatase from Thermus thermophilus HB8.
Biochemistry, 46, 2007
|
|
1IDQ
 
 | |
2DFP
 
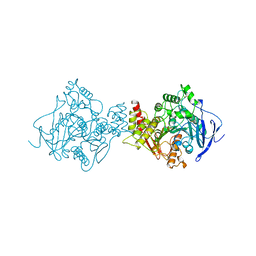 | | X-RAY STRUCTURE OF AGED DI-ISOPROPYL-PHOSPHORO-FLUORIDATE (DFP) BOUND TO ACETYLCHOLINESTERASE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kryger, G, Millard, C.B, Silman, I, Sussman, J.L. | | Deposit date: | 1998-12-07 | | Release date: | 1999-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of aged phosphonylated acetylcholinesterase: nerve agent reaction products at the atomic level.
Biochemistry, 38, 1999
|
|
3BY2
 
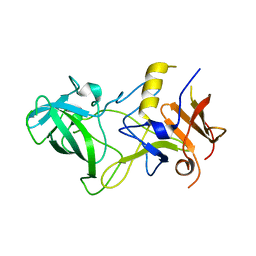 | | Norwalk P polypeptide (228-523) | | Descriptor: | 58 kd capsid protein | | Authors: | Hegde, R, Bu, W. | | Deposit date: | 2008-01-15 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the receptor binding specificity of Norwalk virus.
J.Virol., 82, 2008
|
|
2FFW
 
 | | Solution structure of the RBCC/TRIM B-box1 domain of human MID1: B-box with a RING | | Descriptor: | Midline-1, ZINC ION | | Authors: | Massiah, M.A, Simmons, B.N, Short, K.M, Cox, T.C. | | Deposit date: | 2005-12-20 | | Release date: | 2006-05-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the RBCC/TRIM B-box1 Domain of Human MID1: B-box with a RING.
J.Mol.Biol., 358, 2006
|
|
4E2S
 
 | |
4WNU
 
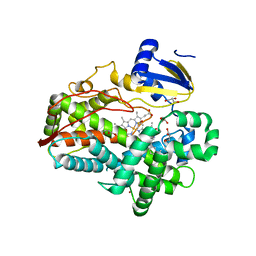 | | Human Cytochrome P450 2D6 Quinidine Complex | | Descriptor: | Cytochrome P450 2D6, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
8PFR
 
 | | Mouse RPL39L integrated into the yeast 60S ribosomal subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Ribosomal protein RPL39L is an efficiency factor in the cotranslational folding of a subset of proteins with alpha helical domains.
Nucleic Acids Res., 52, 2024
|
|
2LAS
 
 | | Molecular Determinants of Paralogue-Specific SUMO-SIM Recognition | | Descriptor: | M-IR2_peptide, Small ubiquitin-related modifier 1 | | Authors: | Namanja, A, Li, Y, Su, Y, Wong, S, Lu, J, Colson, L, Wu, C, Li, S, Chen, Y. | | Deposit date: | 2011-03-20 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into High Affinity Small Ubiquitin-like Modifier (SUMO) Recognition by SUMO-interacting Motifs (SIMs) Revealed by a Combination of NMR and Peptide Array Analysis.
J.Biol.Chem., 287, 2012
|
|
8P8M
 
 | | Yeast 60S ribosomal subunit, RPL39 deletion | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-01 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Ribosomal protein RPL39L is an efficiency factor in the cotranslational folding of a subset of proteins with alpha helical domains.
Nucleic Acids Res., 52, 2024
|
|
7TZW
 
 | |
7TZX
 
 | | The crystal structure of WT CYP199A4 bound to 4-chloromethylbenzoic acid | | Descriptor: | 4-(chloromethyl)benzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Podgorski, M.N, Scaffidi-Muta, J, Bell, S.G. | | Deposit date: | 2022-02-16 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.414 Å) | | Cite: | Cytochrome P450-catalyzed oxidation of halogen-containing substrates.
J.Inorg.Biochem., 244, 2023
|
|
7U00
 
 | |
8VL9
 
 | | Crystal structure of EloBC-VHL-CDO1 complex bound to compound 8 molecular glue | | Descriptor: | CITRIC ACID, Cysteine dioxygenase type 1, Elongin-B, ... | | Authors: | Shu, W, Ma, X, Tutter, A, Buckley, D, Golosov, A, Michaud, G. | | Deposit date: | 2024-01-11 | | Release date: | 2024-03-13 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A small-molecule VHL molecular glue degrader for cysteine dioxygenase 1.
Nat.Chem.Biol., 2025
|
|
8P8U
 
 | | Yeast 60S ribosomal subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Ribosomal protein RPL39L is an efficiency factor in the cotranslational folding of a subset of proteins with alpha helical domains.
Nucleic Acids Res., 52, 2024
|
|
8P8N
 
 | | Mouse RPL39 integrated into the yeast 60S ribosomal subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Ribosomal protein RPL39L is an efficiency factor in the cotranslational folding of a subset of proteins with alpha helical domains.
Nucleic Acids Res., 52, 2024
|
|
6D05
 
 | | Cryo-EM structure of a Plasmodium vivax invasion complex essential for entry into human reticulocytes; two molecules of parasite ligand, subclass 2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Gruszczyk, J, Huang, R.K, Hong, C, Yu, Z, Tham, W.H. | | Deposit date: | 2018-04-10 | | Release date: | 2018-06-20 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of an essential Plasmodium vivax invasion complex.
Nature, 559, 2018
|
|
1TW4
 
 | | Crystal Structure of Chicken Liver Basic Fatty Acid Binding Protein (Bile Acid Binding Protein) Complexed With Cholic Acid | | Descriptor: | CHOLIC ACID, Fatty acid-binding protein | | Authors: | Nichesola, D, Perduca, M, Capaldi, S, Carrizo, M.E, Righetti, P.G, Monaco, H.L. | | Deposit date: | 2004-06-30 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of chicken liver basic Fatty Acid-binding protein complexed with cholic acid
Biochemistry, 43, 2004
|
|
6JG2
 
 | | Crystal structure of barley exohydrolaseI wildtype in complex with 4'-nitrophenyl thiolaminaribioside | | Descriptor: | (2~{R},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-[(2~{S},3~{S},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-(4-nitrophenoxy)-3,5-bis(oxidanyl)oxan-4-yl]sulfanyl-oxane-3,4,5-triol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Barley exohydrolase I, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
3KGC
 
 | | Isolated ligand binding domain dimer of GluA2 ionotropic glutamate receptor in complex with glutamate, LY 404187 and ZK 200775 | | Descriptor: | GLUTAMIC ACID, Glutamate receptor 2, N-[(2S)-2-(4'-cyanobiphenyl-4-yl)propyl]propane-2-sulfonamide, ... | | Authors: | Sobolevsky, A.I, Rosconi, M.P, Gouaux, E. | | Deposit date: | 2009-10-28 | | Release date: | 2009-12-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor
Nature, 462, 2009
|
|
