4Q87
 
 | |
4Q8Y
 
 | |
5YNW
 
 | | Crystal structure of an aromatic prenyltransferase FAMD1 from Fischerella ambigua UTEX 1903 in complex with DMASPP and INN | | Descriptor: | 3-[(Z)-2-isocyanoethenyl]-1H-indole, DIMETHYLALLYL S-THIOLODIPHOSPHATE, GLYCEROL, ... | | Authors: | Wang, J, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-10-25 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insight into a novel indole prenyltransferase in hapalindole-type alkaloid biosynthesis.
Biochem. Biophys. Res. Commun., 495, 2018
|
|
4PZ5
 
 | |
4XAI
 
 | | Crystal Structure of red flour beetle NR2E1/TLX | | Descriptor: | Grunge, isoform J, Maltose-binding periplasmic protein,Tailless ortholog, ... | | Authors: | Zhi, X, Zhou, E, Xu, E. | | Deposit date: | 2014-12-14 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for corepressor assembly by the orphan nuclear receptor TLX.
Genes Dev., 29, 2015
|
|
4Q7V
 
 | |
4Q83
 
 | |
8P8M
 
 | | Yeast 60S ribosomal subunit, RPL39 deletion | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-01 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Ribosomal protein RPL39L is an efficiency factor in the cotranslational folding of a subset of proteins with alpha helical domains.
Nucleic Acids Res., 52, 2024
|
|
4Q8X
 
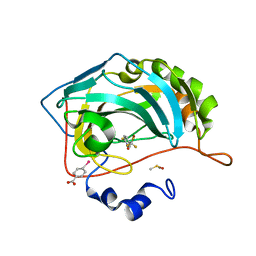 | |
8PFR
 
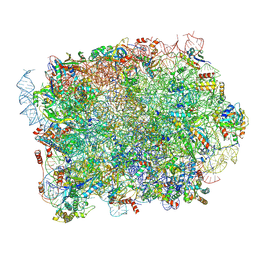 | | Mouse RPL39L integrated into the yeast 60S ribosomal subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Ribosomal protein RPL39L is an efficiency factor in the cotranslational folding of a subset of proteins with alpha helical domains.
Nucleic Acids Res., 52, 2024
|
|
4Q99
 
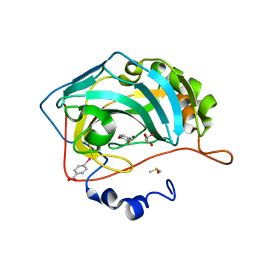 | |
8P8U
 
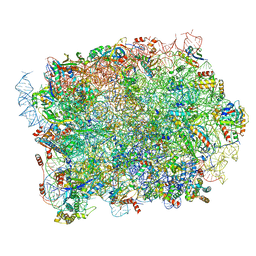 | | Yeast 60S ribosomal subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Ribosomal protein RPL39L is an efficiency factor in the cotranslational folding of a subset of proteins with alpha helical domains.
Nucleic Acids Res., 52, 2024
|
|
8P8N
 
 | | Mouse RPL39 integrated into the yeast 60S ribosomal subunit | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Rabl, J, Banerjee, A, Boehringer, D, Zavolan, M. | | Deposit date: | 2023-06-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Ribosomal protein RPL39L is an efficiency factor in the cotranslational folding of a subset of proteins with alpha helical domains.
Nucleic Acids Res., 52, 2024
|
|
6FNC
 
 | |
3KW0
 
 | |
8Y4Z
 
 | | Monomeric HERC5 HECT c-lobe structure in solution | | Descriptor: | E3 ISG15--protein ligase HERC5 | | Authors: | Dag, C, Lambert, M, Kahraman, K, Lohn, F, Lee, W, Gocenler, O, Guntert, P, Dotsch, V. | | Deposit date: | 2024-01-31 | | Release date: | 2024-02-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Monomeric HERC5 HECT c-lobe structure in solution
To Be Published
|
|
6FN9
 
 | |
3HPI
 
 | |
7SCQ
 
 | |
7SC6
 
 | |
6IY6
 
 | | Crystal structure of human cytosolic aspartyl-tRNA synthetase (DRS) in complex with glutathion-S transferase (GST) domains from Aminoacyl tRNA synthase complex-interacting multifunctional protein 2 (AIMP2) and glutamyl-prolyl-tRNA synthetase (EPRS) | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 2, Aspartate--tRNA ligase, cytoplasmic, ... | | Authors: | Park, S.H, Hahn, H, Han, B.W. | | Deposit date: | 2018-12-13 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The DRS-AIMP2-EPRS subcomplex acts as a pivot in the multi-tRNA synthetase complex.
Iucrj, 6, 2019
|
|
3I5D
 
 | | Crystal structure of the ATP-gated P2X4 ion channel in the closed, apo state at 3.5 Angstroms (R3) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, P2X purinoceptor | | Authors: | Kawate, T, Michel, J.C, Gouaux, E. | | Deposit date: | 2009-07-05 | | Release date: | 2009-08-04 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Crystal structure of the ATP-gated P2X(4) ion channel in the closed state.
Nature, 460, 2009
|
|
3DKS
 
 | | DsbA substrate complex | | Descriptor: | Thiol:disulfide interchange protein dsbA, siga peptide | | Authors: | Paxman, J.J, Borg, N.A, Horne, J, Rossjohn, J, Thompson, P.E, Piek, S, Kahler, C.M, Sakellaris, H, Scanlon, M.J. | | Deposit date: | 2008-06-25 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of the bacterial oxidoreductase enzyme DsbA in complex with a peptide reveals a basis for substrate specificity in the catalytic cycle of DsbA enzymes
J.Biol.Chem., 284, 2009
|
|
6PRF
 
 | | HIV-1 Protease multiple drug resistant clinical isolate mutant PR20 with GRL-14213A | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Protease, YTTRIUM ION | | Authors: | Kneller, D.W, Agniswamy, J, Weber, I.T. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor combats highly drug resistant mutant PR20.
Biochem.Biophys.Res.Commun., 519, 2019
|
|
3DN5
 
 | | Aldose Reductase in complex with novel biarylic inhibitor | | Descriptor: | 3-[5-(3-nitrophenyl)thiophen-2-yl]propanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Klebe, G. | | Deposit date: | 2008-07-01 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Based Optimization of Aldose Reductase Inhibitors Originating from Virtual Screening
Chemmedchem, 4, 2009
|
|
