7RNM
 
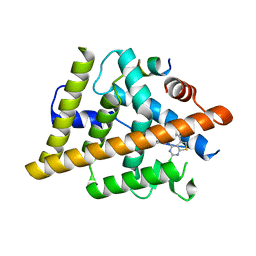 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with 2-(2-Chloro-5-phenylthieno[2,3-d]pyrimidin-4-yl)isoindolin-5-ol and GRIP Peptide | | Descriptor: | 2-(2-chloro-5-phenylthieno[2,3-d]pyrimidin-4-yl)-2,3-dihydro-1H-isoindol-5-ol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Joiner, C, Sammeta, V.K.R, Norris, J.D, McDonnell, D.P, Willson, T.M, Fanning, S.W. | | Deposit date: | 2021-07-29 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Determinants of the Binding and Activation of Estrogen Receptor alpha by Phenolic Thieno[2,3-d]pyrimidines
Helv.Chim.Acta, 106, 2023
|
|
4X04
 
 | | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM CITROBACTER KOSERI (CKO_04899, TARGET EFI-510094) WITH BOUND D-glucuronate | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, beta-D-glucopyranuronic acid, ... | | Authors: | Yadava, U, Vetting, M.W, Al Obaidi, N.F, Toro, R, Morisco, L.L, Benach, J, Wasserman, S.R, Attonito, J.D, Scott Glenn, A, Chamala, S, Chowdhury, S, Lafleur, J, Love, J, Seidel, R.D, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-11-20 | | Release date: | 2014-12-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CRYSTAL STRUCTURE OF A TRAP PERIPLASMIC SOLUTE BINDING PROTEIN FROM CITROBACTER KOSERI (CKO_04899, TARGET EFI-510094) WITH BOUND D-glucuronate
To be published
|
|
4YI3
 
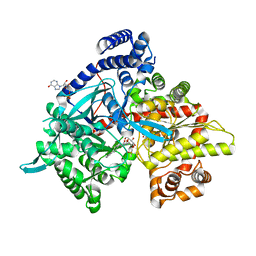 | | Crystal structure of Gpb in complex with 4a | | Descriptor: | DIMETHYL SULFOXIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Kantsadi, A.L, Chatzileontiadou, D.S.M, Stravodimos, G.A, Leonidas, D.D. | | Deposit date: | 2015-02-27 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glycogen phosphorylase as a target for type 2 diabetes: synthetic, biochemical, structural and computational evaluation of novel N-acyl-N -( beta-D-glucopyranosyl) urea inhibitors.
Curr Top Med Chem, 15, 2015
|
|
1I5V
 
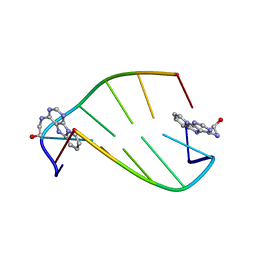 | | SOLUTION STRUCTURE OF 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL INTERCALATED IN THE DNA DUPLEX D(CGATCG)2 | | Descriptor: | 2-(PYRIDO[1,2-E]PURIN-4-YL)AMINO-ETHANOL, 5'-D(*CP*GP*AP*TP*CP*G)-3' | | Authors: | Favier, A, Blackledge, M, Simorre, J.P, Marion, D, Debousy, J.C. | | Deposit date: | 2001-03-01 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 2-(pyrido[1,2-e]purin-4-yl)amino-ethanol intercalated in the DNA duplex d(CGATCG)2.
Biochemistry, 40, 2001
|
|
7SA3
 
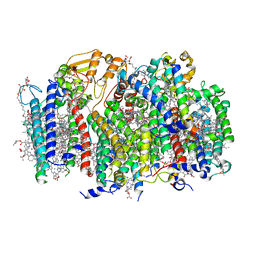 | | Structure of a monomeric photosystem II core complex from a cyanobacterium acclimated to far-red light | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Gisriel, C.J, Bryant, D.A, Brudvig, G.W. | | Deposit date: | 2021-09-22 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Structure of a monomeric photosystem II core complex from a cyanobacterium acclimated to far-red light reveals the functions of chlorophylls d and f.
J.Biol.Chem., 298, 2021
|
|
7C04
 
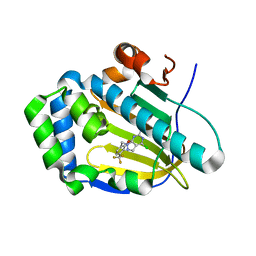 | | Crystal structure of human Trap1 with DN203492 | | Descriptor: | 4-chloranyl-1-[[2-methoxy-4-(trifluoromethyl)phenyl]methyl]pyrazolo[3,4-d]pyrimidin-6-amine, Heat shock protein 75 kDa, mitochondrial | | Authors: | Kim, D, Kim, D, Kim, S.Y, Lee, J.H, Kang, B.H, Kang, S, Lee, C. | | Deposit date: | 2020-04-30 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Development of pyrazolo[3,4-d]pyrimidine-6-amine-based TRAP1 inhibitors that demonstrate in vivo anticancer activity in mouse xenograft models.
Bioorg.Chem., 101, 2020
|
|
7C05
 
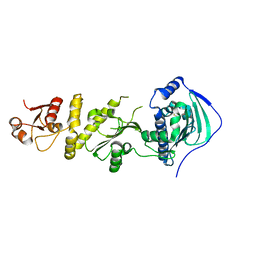 | | Crystal structure of human Trap1 with DN203495 | | Descriptor: | 1-[(4-bromanyl-2-fluoranyl-phenyl)methyl]-4-chloranyl-pyrazolo[3,4-d]pyrimidin-6-amine, Heat shock protein 75 kDa, mitochondrial | | Authors: | Kim, D, Kim, D, Kim, S.Y, Lee, J.H, Kang, B.H, Kang, S, Lee, C. | | Deposit date: | 2020-04-30 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Development of pyrazolo[3,4-d]pyrimidine-6-amine-based TRAP1 inhibitors that demonstrate in vivo anticancer activity in mouse xenograft models.
Bioorg.Chem., 101, 2020
|
|
8R1X
 
 | |
1JPD
 
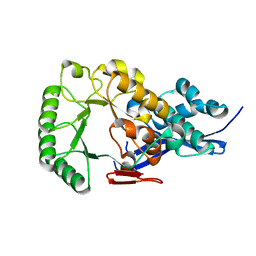 | | L-Ala-D/L-Glu Epimerase | | Descriptor: | L-Ala-D/L-Glu Epimerase | | Authors: | Gulick, A.M, Schmidt, D.M.Z, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-08-01 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystal structures of the L-Ala-D/L-Glu epimerases from Escherichia coli and Bacillus subtilis.
Biochemistry, 40, 2001
|
|
9HTA
 
 | | Dispersin from Terribacillus saccharophilus Dispts2 in complex with NAG-thiazoline | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Dispersin | | Authors: | Males, A, Moroz, O.V, Blagova, E, Munch, A, Hansen, G.H, Johansen, A.H, Ostergaard, L.H, Segura, D.R, Eddenden, A, Due, A.V, Gudmand, M, Salomon, J, Sorensen, S.R, Franco Cairo, J.P.L, Pache, R.A, Vejborg, R.M, Bhosale, S, Vocadlo, D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2024-12-19 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Expansion of the diversity of dispersin scaffolds.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
186D
 
 | |
7AD3
 
 | | Class D GPCR Ste2 dimer coupled to two G proteins | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-factor mating pheromone, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Velazhahan, V, Tate, C. | | Deposit date: | 2020-09-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the class D GPCR Ste2 dimer coupled to two G proteins.
Nature, 589, 2021
|
|
8P3A
 
 | | bacteriophage T5 l-alanoyl-d-glutamate peptidase Zn2+/Ca2+ form | | Descriptor: | CALCIUM ION, L-alanyl-D-glutamate peptidase, ZINC ION | | Authors: | Prokhorov, D.A, Kutyshenko, V.P, Mikoulinskaia, G.V. | | Deposit date: | 2023-05-17 | | Release date: | 2023-07-05 | | Last modified: | 2025-01-15 | | Method: | SOLUTION NMR | | Cite: | Conservative Tryptophan Residue in the Vicinity of an Active Site of the M15 Family l,d-Peptidases: A Key Element in the Catalysis.
Int J Mol Sci, 24, 2023
|
|
6OJH
 
 | | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with (R)-7-(3-aminopyrrolidin-1-yl)-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine | | Descriptor: | 7-[(3R)-3-aminopyrrolidin-1-yl]-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine, ACETATE ION, Biotin carboxylase, ... | | Authors: | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | Deposit date: | 2019-04-11 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Haemophilus Influenzae Biotin Carboxylase Complexed with (R)-7-(3-aminopyrrolidin-1-yl)-6-(naphthalen-1-yl)pyrido[2,3-d]pyrimidin-2-amine
To Be Published
|
|
8XIV
 
 | | Production of D-psicose | | Descriptor: | COBALT (II) ION, D-fructose, D-tagatose 3-epimerase, ... | | Authors: | Guo, D. | | Deposit date: | 2023-12-20 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Production of D-psicose
To Be Published
|
|
1HMA
 
 | | THE SOLUTION STRUCTURE AND DYNAMICS OF THE DNA BINDING DOMAIN OF HMG-D FROM DROSOPHILA MELANOGASTER | | Descriptor: | HMG-D | | Authors: | Jones, D.N.M, Searles, M.A, Shaw, G.L, Churchill, M.E.A, Ner, S.S, Keeler, J, Travers, A.A, Neuhaus, D. | | Deposit date: | 1994-05-12 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of the DNA-binding domain of HMG-D from Drosophila melanogaster.
Structure, 2, 1994
|
|
1D93
 
 | |
1IHH
 
 | | 2.4 ANGSTROM CRYSTAL STRUCTURE OF AN OXALIPLATIN 1,2-D(GPG) INTRASTRAND CROSS-LINK IN A DNA DODECAMER DUPLEX | | Descriptor: | 1R,2R-DIAMINOCYCLOHEXANE, 5'-D(*CP*CP*TP*CP*TP*GP*GP*TP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*AP*CP*CP*AP*GP*AP*GP*G)-3', ... | | Authors: | Spingler, B, Whittington, D.A, Lippard, S.J. | | Deposit date: | 2001-04-19 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2.4 A crystal structure of an oxaliplatin 1,2-d(GpG) intrastrand cross-link in a DNA dodecamer duplex.
Inorg.Chem., 40, 2001
|
|
6QMT
 
 | | Complement factor D in complex with the inhibitor 2-(2-(3'-(aminomethyl)-[1,1'-biphenyl]-3-carboxamido)phenyl)acetic acid | | Descriptor: | 2-[2-[[3-[3-(aminomethyl)phenyl]phenyl]carbonylamino]phenyl]ethanoic acid, Complement factor D | | Authors: | Karki, R, Powers, J, Mainolfi, N, Anderson, K, Belanger, D, Liu, D, Jendza, K, Gelin, C.F, Solovay, C, Mac Sweeeny, A, Delgado, O, Crowley, M, Liao, S.-M, Argikar, U.A, Flohr, S, La Bonte, L.R, Lorthiois, E.L, Vulpetti, A, Cumin, F, Brown, A, Adams, C, Jaffee, B, Mogi, M. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Preclinical Characterization of Selective Factor D Inhibitors Targeting the Alternative Complement Pathway.
J.Med.Chem., 62, 2019
|
|
9GCJ
 
 | |
1JPM
 
 | | L-Ala-D/L-Glu Epimerase | | Descriptor: | GLYCEROL, L-Ala-D/L-Glu Epimerase, MAGNESIUM ION | | Authors: | Gulick, A.M, Schmidt, D.M.Z, Gerlt, J.A, Rayment, I. | | Deposit date: | 2001-08-02 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: crystal structures of the L-Ala-D/L-Glu epimerases from Escherichia coli and Bacillus subtilis.
Biochemistry, 40, 2001
|
|
6QMR
 
 | | Complement factor D in complex with the inhibitor (S)-2-(2-((3'-(1-amino-2-hydroxyethyl)-[1,1'-biphenyl]-3-yl)methoxy)phenyl)acetic acid | | Descriptor: | 2-[2-[[3-[3-[(1~{S})-1-azanyl-2-oxidanyl-ethyl]phenyl]phenyl]methoxy]phenyl]ethanoic acid, Complement factor D | | Authors: | Karki, R, Powers, J, Mainolfi, N, Anderson, K, Belanger, D, Liu, D, Jendza, K, Gelin, C.F, Solovay, C, Mac Sweeeny, A, Delgado, O, Crowley, M, Liao, S.-M, Argikar, U.A, Flohr, S, La Bonte, L.R, Lorthiois, E.L, Vulpetti, A, Cumin, F, Brown, A, Adams, C, Jaffee, B, Mogi, M. | | Deposit date: | 2019-02-08 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, Synthesis, and Preclinical Characterization of Selective Factor D Inhibitors Targeting the Alternative Complement Pathway.
J.Med.Chem., 62, 2019
|
|
1KNK
 
 | | Crystal Structure of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate Synthase (ispF) from E. coli involved in Mevalonate-Independent Isoprenoid Biosynthesis | | Descriptor: | 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase, MANGANESE (II) ION | | Authors: | Richard, S.B, Ferrer, J.L, Bowman, M.E, Lillo, A.M, Tetzlaff, C.N, Cane, D.E, Noel, J.P. | | Deposit date: | 2001-12-18 | | Release date: | 2002-06-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase. An enzyme in the mevalonate-independent isoprenoid biosynthetic pathway.
J.Biol.Chem., 277, 2002
|
|
1D91
 
 | |
5TOL
 
 | | CRYSTAL STRUCTURE OF BETA-SITE APP-CLEAVING ENZYME 1 COMPLEXED WITH N-(3-((4AS,7AS)-2-AMINO-4,4A,5,6-TETRAHYDRO-7AH-FURO[2,3-D][1,3]THIAZIN-7A-YL)-4-FLUOROPHENYL)-5-BROMO-2-PYRIDINECARBOXAMIDE | | Descriptor: | Beta-secretase 1, N-{3-[(4aR,7aR)-2-amino-4,4a,5,6-tetrahydro-7aH-furo[2,3-d][1,3]thiazin-7a-yl]-4-fluorophenyl}-5-bromopyridine-2-carboxamide | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2016-10-18 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of furo[2,3-d][1,3]thiazinamines as beta amyloid cleaving enzyme-1 (BACE1) inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
