4F1Z
 
 | | Crystal structures reveal the multi-ligand binding mechanism of the Staphylococcus aureus ClfB | | Descriptor: | Clumping factor B, MAGNESIUM ION, peptide from Keratin, ... | | Authors: | Yang, M.J, Xiang, H, Wang, J.W, Liu, B, Chen, Y.G, Liu, L, Deng, X.M, Feng, Y. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures Reveal the Multi-Ligand Binding Mechanism of Staphylococcus aureus ClfB
Plos Pathog., 8, 2012
|
|
4F24
 
 | | Crystal structures reveal the multi-ligand binding mechanism of the Staphylococcus aureus ClfB | | Descriptor: | Clumping factor B, MAGNESIUM ION | | Authors: | Yang, M.J, Xiang, H, Wang, J.W, Liu, B, Chen, Y.G, Liu, L, Deng, X.M, Feng, Y. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Crystal Structures Reveal the Multi-Ligand Binding Mechanism of Staphylococcus aureus ClfB
Plos Pathog., 8, 2012
|
|
9FQL
 
 | |
5VVI
 
 | | Crystal Structure of the Ligand Binding Domain of LysR-type Transcriptional Regulator, OccR from Agrobacterium tumefaciens in the Complex with Octopine | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Kim, Y, Chhor, G, Jedrzejczak, R, Winans, S.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-05-19 | | Release date: | 2017-06-21 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Crystal Structure of the Ligand-Binding Domain of a LysR-type Transcriptional Regulator: Transcriptional Activation via a Rotary Switch.
Mol. Microbiol., 2018
|
|
7MK7
 
 | | Augmentor domain of augmentor-beta | | Descriptor: | ALK and LTK ligand 1,Maltodextrin-binding protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Krimmer, S.G, Reshetnyak, A.V, Puleo, D.E, Schlessinger, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.42815185 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
2XNU
 
 | | Acetylcholine binding protein (AChBP) as template for hierarchical in silico screening procedures to identify structurally novel ligands for the nicotinic receptors | | Descriptor: | 2-(2-(4-PHENYLPIPERIDIN-1-YL)ETHYL)-1H-INDOLE, SOLUBLE ACETYLCHOLINE RECEPTOR | | Authors: | Rucktooa, P, Akdemir, A, deEsch, I, Sixma, T.K. | | Deposit date: | 2010-08-06 | | Release date: | 2011-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Acetylcholine Binding Protein (Achbp) as Template for Hierarchical in Silico Screening Procedures to Identify Structurally Novel Ligands for the Nicotinic Receptors.
Bioorg.Med.Chem., 19, 2011
|
|
1NNP
 
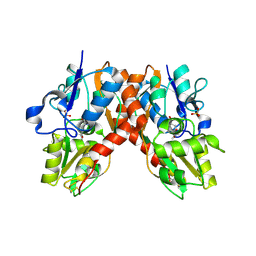 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-ATPA at 1.9 A resolution. Crystallization without zinc ions. | | Descriptor: | 3-(5-TERT-BUTYL-3-OXIDOISOXAZOL-4-YL)-L-ALANINATE, Glutamate receptor 2, SULFATE ION | | Authors: | Lunn, M.L, Hogner, A, Stensbol, T.B, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2003-01-14 | | Release date: | 2003-03-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three-Dimensional Structure of the Ligand-Binding
Core of GluR2 in Complex with the Agonist (S)-ATPA:
Implications for Receptor Subunit Selectivity.
J.Med.Chem., 46, 2003
|
|
7Q3H
 
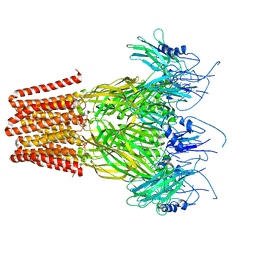 | | Pentameric ligand-gated ion channel, DeCLIC at pH 7 with 10 mM EDTA | | Descriptor: | Neur_chan_LBD domain-containing protein | | Authors: | Lycksell, M, Rovsnik, U, Hanke, A, Howard, R.J, Lindahl, E. | | Deposit date: | 2021-10-27 | | Release date: | 2022-11-16 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Biophysical characterization of calcium-binding and modulatory-domain dynamics in a pentameric ligand-gated ion channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q3G
 
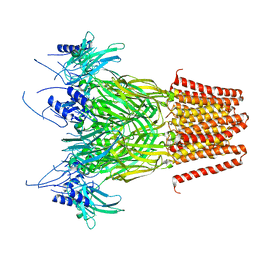 | | Pentameric ligand-gated ion channel, DeCLIC at pH 7 with 10 mM Ca2+ | | Descriptor: | CALCIUM ION, Neur_chan_LBD domain-containing protein | | Authors: | Licksell, M, Rovsnik, U, Hanke, A, Howard, R.J, Lindahl, E. | | Deposit date: | 2021-10-27 | | Release date: | 2022-11-16 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Biophysical characterization of calcium-binding and modulatory-domain dynamics in a pentameric ligand-gated ion channel.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1NNK
 
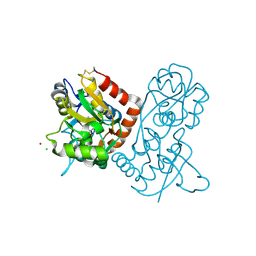 | | X-ray structure of the GluR2 ligand-binding core (S1S2J) in complex with (S)-ATPA at 1.85 A resolution. Crystallization with zinc ions. | | Descriptor: | 3-(5-TERT-BUTYL-3-OXIDOISOXAZOL-4-YL)-L-ALANINATE, CHLORIDE ION, Glutamate receptor 2, ... | | Authors: | Lunn, M.-L, Hogner, A, Stensbol, T.B, Gouaux, E, Egebjerg, J, Kastrup, J.S. | | Deposit date: | 2003-01-14 | | Release date: | 2003-03-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Three-Dimensional Structure of the Ligand-Binding
Core of GluR2 in Complex with the Agonist (S)-ATPA:
Implications for Receptor Subunit Selectivity.
J.Med.Chem., 46, 2003
|
|
3F1O
 
 | | Crystal structure of the high affinity heterodimer of HIF2 alpha and ARNT C-terminal PAS domains, with an internally-bound artificial ligand | | Descriptor: | 1,2-ETHANEDIOL, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1, ... | | Authors: | Scheuermann, T.H, Tomchick, D.R, Machius, M, Guo, Y, Bruick, R.K, Gardner, K.H. | | Deposit date: | 2008-10-28 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8G6P
 
 | | Crystal structure of Mycobacterium thermoresistibile MurE in complex with ADP and 2,6-Diaminopimelic acid | | Descriptor: | 2,6-DIAMINOPIMELIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Rossini, N.O, Silva, C.S, Dias, M.V.B. | | Deposit date: | 2023-02-15 | | Release date: | 2023-04-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The crystal structure of Mycobacterium thermoresistibile MurE ligase reveals the binding mode of the substrate m-diaminopimelate.
J.Struct.Biol., 215, 2023
|
|
2G44
 
 | | Human Estrogen Receptor Alpha Ligand-Binding Domain In Complex With OBCP-1M-G and A Glucocorticoid Receptor Interacting Protein 1 NR Box II Peptide | | Descriptor: | 4-[(1S,2R,5S)-4,4,8-TRIMETHYL-3-OXABICYCLO[3.3.1]NON-7-EN-2-YL]PHENOL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Hsieh, R.W, Sharma, S.K, Greene, G.L. | | Deposit date: | 2006-02-21 | | Release date: | 2007-01-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and characterization of novel estrogen
receptor agonist ligands and development of biochips for
nuclear receptor drug discovery
Thesis, 2006
|
|
3HCQ
 
 | | Structural analysis of the choline binding protein ChoX in a semi-closed and ligand-free conformation | | Descriptor: | Putative choline ABC transporter, periplasmic solute-binding component | | Authors: | Oswald, C, Smits, S.H.J, Hoeing, M, Bremer, E, Schmitt, L. | | Deposit date: | 2009-05-06 | | Release date: | 2009-10-13 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural analysis of the choline-binding protein ChoX in a semi-closed and ligand-free conformation.
Biol.Chem., 390, 2009
|
|
3BFU
 
 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (R)-TDPA at 1.95 A resolution | | Descriptor: | (2R)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, Glutamate receptor 2 | | Authors: | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-11-23 | | Release date: | 2008-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
1DO3
 
 | | CARBONMONOXY-MYOGLOBIN (MUTANT L29W) AFTER PHOTOLYSIS AT T>180K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ostermann, A, Waschipky, R, Parak, F.G, Nienhaus, G.U. | | Deposit date: | 1999-12-18 | | Release date: | 2000-01-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Ligand binding and conformational motions in myoglobin.
Nature, 404, 2000
|
|
9J79
 
 | | Cryo-EM structure of CRL2-FEM1B bound with TOM20(tetramer) | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Zhao, S, Xu, C. | | Deposit date: | 2024-08-18 | | Release date: | 2025-04-09 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | TOM20-driven E3 ligase recruitment regulates mitochondrial dynamics through PLD6.
Nat.Chem.Biol., 2025
|
|
3BFT
 
 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (S)-TDPA at 2.25 A resolution | | Descriptor: | (2S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-11-23 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
9J77
 
 | | Cryo-EM structure of CRL2-FEM1B (dimer 1) | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Zhao, S, Xu, C. | | Deposit date: | 2024-08-18 | | Release date: | 2025-04-09 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | TOM20-driven E3 ligase recruitment regulates mitochondrial dynamics through PLD6.
Nat.Chem.Biol., 2025
|
|
9J78
 
 | | Cryo-EM structure of CRL2-FEM1B (dimer 2) | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Zhao, S, Xu, C. | | Deposit date: | 2024-08-18 | | Release date: | 2025-04-09 | | Last modified: | 2025-05-07 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | TOM20-driven E3 ligase recruitment regulates mitochondrial dynamics through PLD6.
Nat.Chem.Biol., 2025
|
|
1DO7
 
 | | CARBONMONOXY-MYOGLOBIN (MUTANT L29W) REBINDING STRUCTURE AFTER PHOTOLYSIS AT T< 180K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ostermann, A, Waschipky, R, Parak, F.G, Nienhaus, G.U. | | Deposit date: | 1999-12-19 | | Release date: | 2000-01-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ligand binding and conformational motions in myoglobin.
Nature, 404, 2000
|
|
1DO4
 
 | | CARBONMONOXY-MYOGLOBIN (MUTANT L29W) AFTER PHOTOLYSIS AT T<180K | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ostermann, A, Waschipky, R, Parak, F.G, Nienhaus, G.U. | | Deposit date: | 1999-12-18 | | Release date: | 2000-01-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand binding and conformational motions in myoglobin.
Nature, 404, 2000
|
|
1MD2
 
 | | CHOLERA TOXIN B-PENTAMER WITH DECAVALENT LIGAND BMSC-0013 | | Descriptor: | 3-ETHYLAMINO-4-METHYLAMINO-CYCLOBUTANE-1,2-DIONE, CHOLERA TOXIN B SUBUNIT, CYANIDE ION, ... | | Authors: | Zhang, Z, Merritt, E.A, Ahn, M, Roach, C, Hol, W.G.J, Fan, E. | | Deposit date: | 2002-08-06 | | Release date: | 2002-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Solution and crystallographic studies of branched multivalent ligands that inhibit the receptor-binding of cholera toxin.
J.Am.Chem.Soc., 124, 2002
|
|
2WEY
 
 | | Human PDE-papaverine complex obtained by ligand soaking of cross- linked protein crystals | | Descriptor: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXYISOQUINOLINE, CAMP AND CAMP-INHIBITED CGMP 3', 5'-CYCLIC PHOSPHODIESTERASE, ... | | Authors: | Andersen, O.A, Schonfeld, D.L, Toogood-Johnson, I, Felicetti, B, Albrecht, C, Fryatt, T, Whittaker, M, Hallett, D, Barker, J. | | Deposit date: | 2009-04-02 | | Release date: | 2009-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cross-Linking of Protein Crystals as an Aid in the Generation of Binary Protein-Ligand Crystal Complexes, Exemplified by the Human Pde10A-Papaverine Structure.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2FAI
 
 | | Human Estrogen Receptor Alpha Ligand-Binding Domain In Complex With OBCP-2M and A Glucocorticoid Receptor Interacting Protein 1 NR Box II Peptide | | Descriptor: | 4-[(1S,2S,5S,9R)-5-(HYDROXYMETHYL)-8,9-DIMETHYL-3-OXABICYCLO[3.3.1]NON-7-EN-2-YL]PHENOL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Hsieh, R.W, Sharma, S.K, Greene, G.L. | | Deposit date: | 2005-12-07 | | Release date: | 2006-05-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of ligands with bicyclic scaffolds provides insights into mechanisms of estrogen receptor subtype selectivity.
J.Biol.Chem., 281, 2006
|
|
