6VCB
 
 | | Cryo-EM structure of the Glucagon-like peptide-1 receptor in complex with G protein, GLP-1 peptide and a positive allosteric modulator | | Descriptor: | 1-[(1R)-1-(2,6-dichloro-3-methoxyphenyl)ethyl]-6-{2-[(2R)-piperidin-2-yl]phenyl}-1H-benzimidazole, Glucagon-like peptide 1, Glucagon-like peptide 1 receptor, ... | | Authors: | Sun, B, Feng, D, Bueno, A, Kobilka, B, Sloop, K. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into probe-dependent positive allosterism of the GLP-1 receptor.
Nat.Chem.Biol., 16, 2020
|
|
1RC4
 
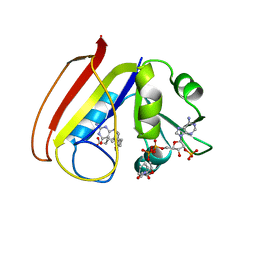 | |
6AGT
 
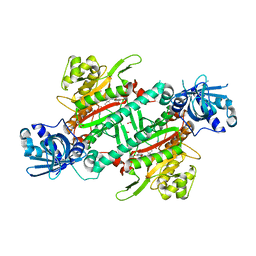 | | Crystal structure of PfKRS complexed with chromone inhibitor | | Descriptor: | COBALT (II) ION, FORMIC ACID, LYSINE, ... | | Authors: | Yogavel, M, Sharma, A, Sharma, A, Baragana, B, Walpole, C. | | Deposit date: | 2018-08-14 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.953 Å) | | Cite: | Lysyl-tRNA synthetase as a drug target in malaria and cryptosporidiosis.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
3QHO
 
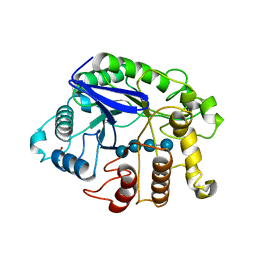 | | Crystal analysis of the complex structure, Y299F-cellotetraose, of endocellulase from pyrococcus horikoshii | | Descriptor: | 458aa long hypothetical endo-1,4-beta-glucanase, PHOSPHATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kim, H.-W, Ishikawa, K. | | Deposit date: | 2011-01-26 | | Release date: | 2012-02-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Functional analysis of hyperthermophilic endocellulase from Pyrococcus horikoshii by crystallographic snapshots
Biochem.J., 437, 2011
|
|
4FCR
 
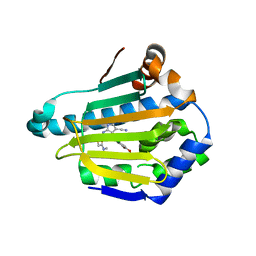 | | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization | | Descriptor: | 2-{[4-(2-chloro-4,5-dimethoxyphenyl)-5-cyano-7H-pyrrolo[2,3-d]pyrimidin-2-yl]sulfanyl}-N,N-dimethylacetamide, Heat shock protein HSP 90-alpha | | Authors: | Davies, N.G, Browne, H, Davis, B, Foloppe, N, Geoffrey, S, Gibbons, B, Hart, T, Drysdale, M.J, Mansell, H, Massey, A, Matassova, N, Moore, J.D, Murray, J, Pratt, R, Ray, S, Roughley, S.D, Jensen, M.R, Schoepfer, J, Scriven, K, Simmonite, H, Stokes, S, Surgenor, A, Webb, P, Wright, L, Brough, P. | | Deposit date: | 2012-05-25 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Targeting conserved water molecules: Design of 4-aryl-5-cyanopyrrolo[2,3-d]pyrimidine Hsp90 inhibitors using fragment-based screening and structure-based optimization.
Bioorg.Med.Chem., 20, 2012
|
|
1RX5
 
 | |
4RR6
 
 | | N-terminal editing domain of threonyl-tRNA synthetase from Aeropyrum pernix with L-Ser3AA (snapshot 1) | | Descriptor: | Probable threonine--tRNA ligase 2, SERINE-3'-AMINOADENOSINE | | Authors: | Ahmad, S, Muthukumar, S, Yerabham, A.S.K, Kamarthapu, V, Sankaranarayanan, R. | | Deposit date: | 2014-11-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Specificity and catalysis hardwired at the RNA-protein interface in a translational proofreading enzyme.
Nat Commun, 6, 2015
|
|
1RX9
 
 | |
4RRQ
 
 | |
3MH8
 
 | |
2G9Y
 
 | |
5IOV
 
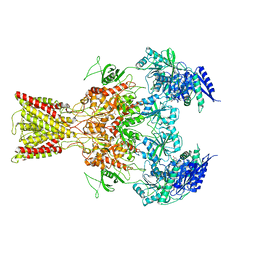 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the glutamate/glycine/Ro25-6981-bound conformation | | Descriptor: | 4-[(1R,2S)-3-(4-benzylpiperidin-1-yl)-1-hydroxy-2-methylpropyl]phenol, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, Mchaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-09 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
5IPV
 
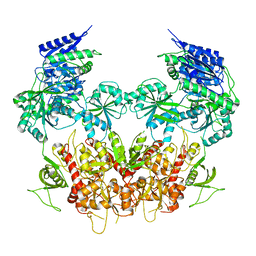 | | Cryo-EM structure of GluN1/GluN2B NMDA receptor in the DCKA/D-APV-bound conformation, state 1 | | Descriptor: | Ionotropic glutamate receptor subunit NR2B, N-methyl-D-aspartate receptor subunit NR1-8a | | Authors: | Zhu, S, Stein, A.R, Yoshioka, C, Lee, C.H, Goehring, A, McHaourab, S.H, Gouaux, E. | | Deposit date: | 2016-03-10 | | Release date: | 2016-04-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (9.25 Å) | | Cite: | Mechanism of NMDA Receptor Inhibition and Activation.
Cell, 165, 2016
|
|
1RX3
 
 | |
5VK0
 
 | |
5A31
 
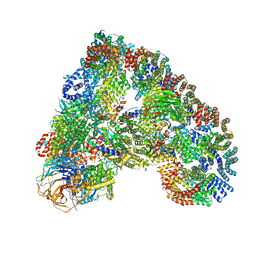 | | Structure of the human APC-Cdh1-Hsl1-UbcH10 complex. | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, Mclaughlin, S.H, Barford, D. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Atomic Structure of the Apc/C and its Mechanism of Protein Ubiquitination.
Nature, 522, 2015
|
|
8OGD
 
 | | Structure of zinc(II) double mutant human carbonic anhydrase II bound to thiocyanate | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase 2, THIOCYANATE ION, ... | | Authors: | Silva, J.M, Cerofolini, L, Carvalho, A.L, Ravera, E, Fragai, M, Parigi, G, Macedo, A.L, Geraldes, C.F.G.C, Luchinat, C. | | Deposit date: | 2023-03-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Elucidating the concentration-dependent effects of thiocyanate binding to carbonic anhydrase.
J.Inorg.Biochem., 244, 2023
|
|
4FSE
 
 | |
8OGE
 
 | | Structure of cobalt(II) substituted double mutant human carbonic anhydrase II bound to thiocyanate | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, COBALT (II) ION, Carbonic anhydrase 2, ... | | Authors: | Silva, J.M, Cerofolini, L, Carvalho, A.L, Ravera, E, Fragai, M, Parigi, G, Macedo, A.L, Geraldes, C.F.G.C, Luchinat, C. | | Deposit date: | 2023-03-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Elucidating the concentration-dependent effects of thiocyanate binding to carbonic anhydrase.
J.Inorg.Biochem., 244, 2023
|
|
4IQ6
 
 | | Gsk-3beta with inhibitor 6-chloro-N-cyclohexyl-4-(1H-pyrrolo[2,3-b]pyridin-3-yl)pyridin-2-amine | | Descriptor: | 6-chloro-N-cyclohexyl-4-(1H-pyrrolo[2,3-b]pyridin-3-yl)pyridin-2-amine, Glycogen synthase kinase-3 beta | | Authors: | Tong, Y, Stewart, K.D, Florjancic, A.S, Harlan, J.E, Merta, P.J, Przytulinska, M, Soni, N, Swinger, K.S, Zhu, H, Johnson, E.F, Shoemaker, A.R, Penning, T.D. | | Deposit date: | 2013-01-10 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Azaindole-Based Inhibitors of Cdc7 Kinase: Impact of the Pre-DFG Residue, Val 195.
ACS Med Chem Lett, 4, 2013
|
|
3GI5
 
 | | Crystal structure of protease inhibitor, KB62 in complex with wild type HIV-1 protease | | Descriptor: | (5S)-3-(3-Acetylphenyl)-N-[(1S,2R)-3-[(1,3-benzodioxol-5-ylsulfonyl)(2-methylpropyl)amino]-2-hydroxy-1-(phenylmethyl)pr opyl]-2-oxo-5-oxazolidinecarboxamide, PHOSPHATE ION, Protease | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2009-03-05 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance.
J.Virol., 84, 2010
|
|
4FL7
 
 | |
4RR7
 
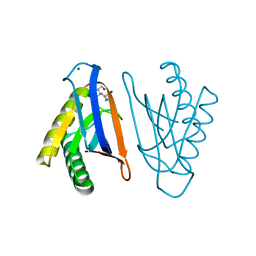 | | N-terminal editing domain of threonyl-tRNA synthetase from Aeropyrum pernix with L-Ser3AA (snapshot 2) | | Descriptor: | MAGNESIUM ION, Probable threonine--tRNA ligase 2, SERINE-3'-AMINOADENOSINE | | Authors: | Ahmad, S, Muthukumar, S, Yerabham, A.S.K, Kamarthapu, V, Sankaranarayanan, R. | | Deposit date: | 2014-11-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Specificity and catalysis hardwired at the RNA-protein interface in a translational proofreading enzyme.
Nat Commun, 6, 2015
|
|
4RRG
 
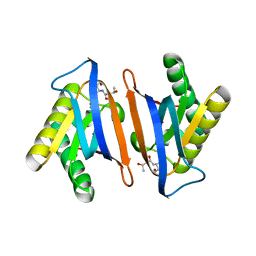 | | Editing domain of threonyl-tRNA synthetase from Methanococcus jannaschii with L-Thr3AA | | Descriptor: | 3'-deoxy-3'-(L-threonylamino)adenosine, Threonine--tRNA ligase | | Authors: | Ahmad, S, Yerabham, A.S.K, Kamarthapu, V, Sankaranarayanan, R. | | Deposit date: | 2014-11-06 | | Release date: | 2015-07-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Specificity and catalysis hardwired at the RNA-protein interface in a translational proofreading enzyme.
Nat Commun, 6, 2015
|
|
7TND
 
 | |
