8U8H
 
 | |
8U8F
 
 | | GPR3 Orphan G-coupled Protein Receptor in complex with Dominant Negative Gs. | | Descriptor: | G-protein coupled receptor 3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Russell, I.C, Belousoff, M.J, Sexton, P. | | Deposit date: | 2023-09-17 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Lipid-Dependent Activation of the Orphan G Protein-Coupled Receptor, GPR3.
Biochemistry, 63, 2024
|
|
8U8B
 
 | | Cryo-EM structure of LRRK2 bound to type II inhibitor rebastinib | | Descriptor: | 4-[4-({[3-tert-butyl-1-(quinolin-6-yl)-1H-pyrazol-5-yl]carbamoyl}amino)-3-fluorophenoxy]-N-methylpyridine-2-carboxamide, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-09-16 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Pharmacology of LRRK2 with type I and II kinase inhibitors revealed by cryo-EM.
Cell Discov, 10, 2024
|
|
8U8A
 
 | | Cryo-EM structure of LRRK2 bound to type II inhibitor ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-09-16 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Pharmacology of LRRK2 with type I and II kinase inhibitors revealed by cryo-EM.
Cell Discov, 10, 2024
|
|
8U89
 
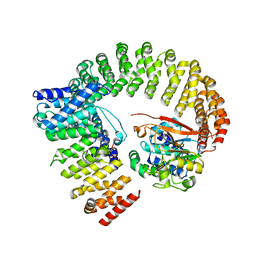 | | The structure of the PP2A-B56Delta holoenzyme mutant - E197K | | Descriptor: | MANGANESE (II) ION, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit delta isoform, Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform, ... | | Authors: | Wu, C.G, Xing, Y. | | Deposit date: | 2023-09-16 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | B56 delta long-disordered arms form a dynamic PP2A regulation interface coupled with global allostery and Jordan's syndrome mutations.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U87
 
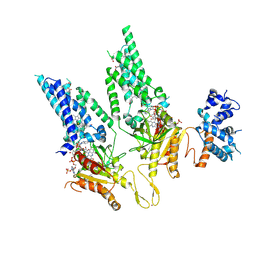 | |
8U86
 
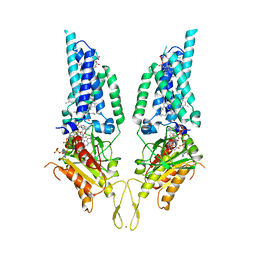 | |
8U85
 
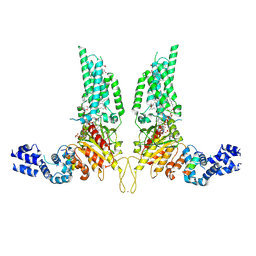 | | Structural Basis of Human NOX5 Activation | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA-ALA, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Cui, C, Jiang, M, Sun, J. | | Deposit date: | 2023-09-15 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Basis of Human NOX5 Activation
To Be Published
|
|
8U84
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State D From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U83
 
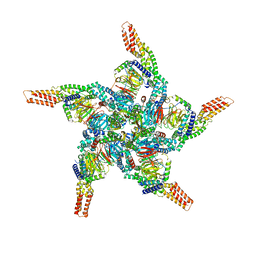 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State C From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.975 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U82
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State B From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U81
 
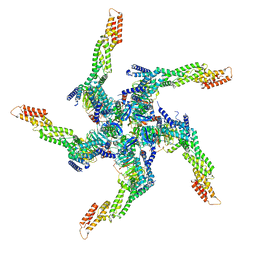 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State A From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U80
 
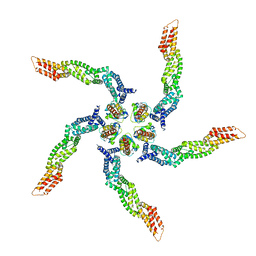 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: Local Refinment of KCTD5(BTB)/Cullin3(NTD) | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3 | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U7Z
 
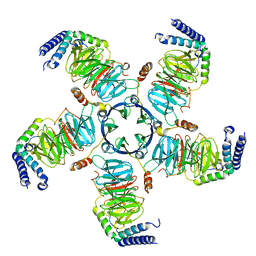 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: Local Refinment of KCTD5(CTD)/Gbeta1gamma2 | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U7Y
 
 | |
8U7X
 
 | |
8U7W
 
 | |
8U7V
 
 | | Human retinal variant phosphomimetic IMPDH1(546)-S477D filament bound by GTP, ATP, IMP, and NAD+, interface-centered | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Calise, S.J, Kollman, J.M. | | Deposit date: | 2023-09-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Light-sensitive phosphorylation regulates retinal IMPDH1 activity and filament assembly.
J.Cell Biol., 223, 2024
|
|
8U7U
 
 | | Proteasome 20S Core Particle from Beta 3 D205 deletion | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Walsh Jr, R.M, Rawson, S, Velez, B, Blickling, M, Razi, A, Hanna, J. | | Deposit date: | 2023-09-15 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Mechanism of autocatalytic activation during proteasome assembly.
Nat.Struct.Mol.Biol., 2024
|
|
8U7Q
 
 | | Human retinal variant phosphomimetic IMPDH1(546)-S477D filament bound by GTP, ATP, IMP, and NAD+, octamer-centered | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Calise, S.J, Kollman, J.M. | | Deposit date: | 2023-09-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Light-sensitive phosphorylation regulates retinal IMPDH1 activity and filament assembly.
J.Cell Biol., 223, 2024
|
|
8U7M
 
 | | Human retinal variant phosphomimetic IMPDH1(595)-S477D free octamer bound by GTP, ATP, IMP, and NAD+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Calise, S.J, Kollman, J.M. | | Deposit date: | 2023-09-15 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Light-sensitive phosphorylation regulates retinal IMPDH1 activity and filament assembly.
J.Cell Biol., 223, 2024
|
|
8U7L
 
 | | Cryo-EM structure of LRRK2 bound to type II inhibitor GZD824 | | Descriptor: | 4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}-3-[(1H-pyrazolo[3,4-b]pyridin-5-yl)ethynyl]benzamide, GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2 | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-09-15 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Pharmacology of LRRK2 with type I and II kinase inhibitors revealed by cryo-EM.
Cell Discov, 10, 2024
|
|
8U7I
 
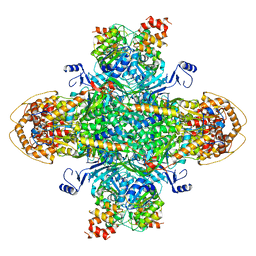 | | Structure of the phage immune evasion protein Gad1 bound to the Gabija GajAB complex | | Descriptor: | Endonuclease GajA, Gabija Anti-Defense 1, Gabija protein GajB | | Authors: | Antine, S.P, Johnson, A.G, Mooney, S.E, Mayer, M.L, Kranzusch, P.J. | | Deposit date: | 2023-09-15 | | Release date: | 2023-11-22 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis of Gabija anti-phage defence and viral immune evasion.
Nature, 625, 2024
|
|
8U7H
 
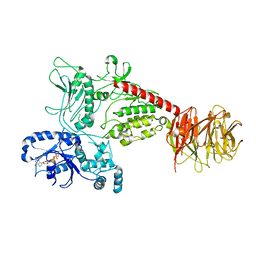 | | Cryo-EM structure of LRRK2 bound to type I inhibitor GNE-7915 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, [4-[[4-(ethylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino]-2-fluoranyl-5-methoxy-phenyl]-morpholin-4-yl-methanone, non-specific serine/threonine protein kinase | | Authors: | Zhu, H, Sun, J. | | Deposit date: | 2023-09-15 | | Release date: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Pharmacology of LRRK2 with type I and II kinase inhibitors revealed by cryo-EM.
Cell Discov, 10, 2024
|
|
8U7E
 
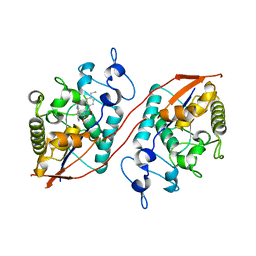 | | Structure of Sts-1 HP domain with rebamipide derivative | | Descriptor: | N-(4-ethylbenzoyl)-3-(2-oxo-1,2-dihydroquinolin-4-yl)-L-alanine, Ubiquitin-associated and SH3 domain-containing protein B | | Authors: | Aziz, F, Dey, R, French, J.B. | | Deposit date: | 2023-09-15 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Rebamipide and Derivatives are Potent, Selective Inhibitors of Histidine Phosphatase Activity of the Suppressor of T Cell Receptor Signaling Proteins.
J.Med.Chem., 67, 2024
|
|
