7P3Q
 
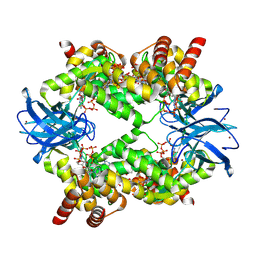 | | Streptomyces coelicolor dATP/ATP-loaded NrdR octamer | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Transcriptional repressor NrdR, ... | | Authors: | Martinez-Carranza, M, Stenmark, P. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-11 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | A nucleotide-sensing oligomerization mechanism that controls NrdR-dependent transcription of ribonucleotide reductases.
Nat Commun, 13, 2022
|
|
7P3F
 
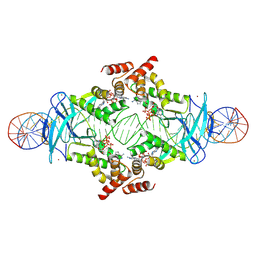 | |
7P37
 
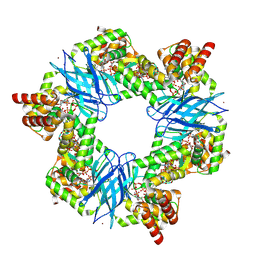 | | Streptomyces coelicolor ATP-loaded NrdR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transcriptional repressor NrdR, ZINC ION | | Authors: | Martinez-Carranza, M, Stenmark, P. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-11 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | A nucleotide-sensing oligomerization mechanism that controls NrdR-dependent transcription of ribonucleotide reductases.
Nat Commun, 13, 2022
|
|
8P23
 
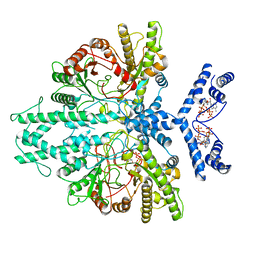 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, ATP/CTP-bound state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-14 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
Elife, 2023
|
|
8P27
 
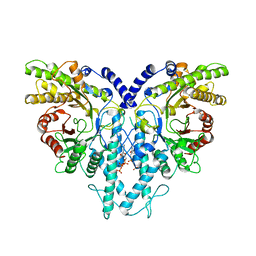 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, dATP-bound state | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, MAGNESIUM ION | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductases: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
Elife, 2023
|
|
8P28
 
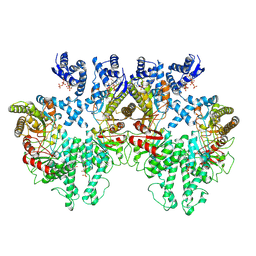 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its tetrameric, dATP-bound state | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, MAGNESIUM ION | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
To Be Published
|
|
8P2D
 
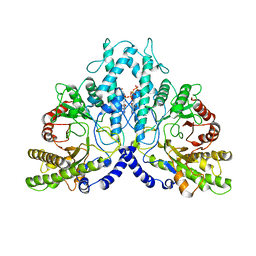 | | Cryo-EM structure of the dimeric form of the anaerobic ribonucleotide reductase from Prevotella copri produced in the presence of dATP and CTP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, MAGNESIUM ION | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
eLife, 2023
|
|
8P2C
 
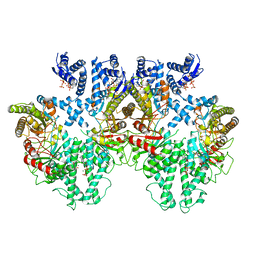 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its tetrameric state produced in the presence of dATP and CTP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, MAGNESIUM ION | | Authors: | Banerjee, I, Bimai, O, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
eLife, 2023
|
|
8P2S
 
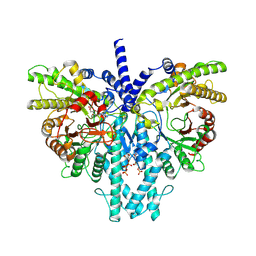 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, ATP/dTTP/GTP-bound state | | Descriptor: | Anaerobic ribonucleoside-triphosphate reductase, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bimai, O, Banerjee, I, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-16 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
eLife, 2023
|
|
8P39
 
 | | Cryo-EM structure of the anaerobic ribonucleotide reductase from Prevotella copri in its dimeric, dGTP/ATP-bound state | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Anaerobic ribonucleoside-triphosphate reductase, ... | | Authors: | Bimai, O, Banerjee, I, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2023-05-17 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Activity modulation in anaerobic ribonucleotide reductase: nucleotide binding to the ATP-cone mediates long-range order-disorder transitions in the active site
To be published
|
|
7R1R
 
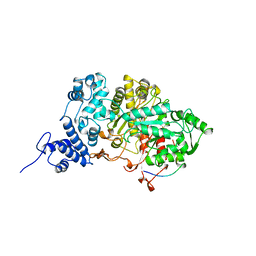 | | RIBONUCLEOTIDE REDUCTASE E441Q MUTANT R1 PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | RIBONUCLEOTIDE REDUCTASE R1 PROTEIN, RIBONUCLEOTIDE REDUCTASE R2 PROTEIN | | Authors: | Eriksson, M, Eklund, H. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase.
J.Biol.Chem., 272, 1997
|
|
6W4X
 
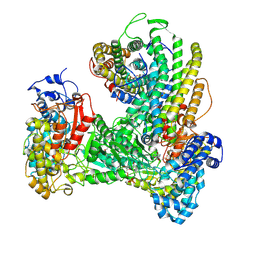 | | Holocomplex of E. coli class Ia ribonucleotide reductase with GDP and TTP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MU-OXO-DIIRON, ... | | Authors: | Kang, G, Taguchi, A, Stubbe, J, Drennan, C. | | Deposit date: | 2020-03-11 | | Release date: | 2020-04-08 | | Last modified: | 2020-05-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a trapped radical transfer pathway within a ribonucleotide reductase holocomplex.
Science, 368, 2020
|
|
6AUI
 
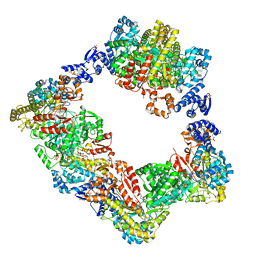 | | Human ribonucleotide reductase large subunit (alpha) with dATP and CDP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Brignole, E.J, Drennan, C.L, Asturias, F.J, Tsai, K.L, Penczek, P.A. | | Deposit date: | 2017-09-01 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | 3.3- angstrom resolution cryo-EM structure of human ribonucleotide reductase with substrate and allosteric regulators bound.
Elife, 7, 2018
|
|
4X3V
 
 | | Crystal structure of human ribonucleotide reductase 1 bound to inhibitor | | Descriptor: | N~6~-{N-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)acetyl]-2-methyl-D-alanyl}-D-lysine, Ribonucleoside-diphosphate reductase large subunit, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Dealwis, C.G, Ahmad, M.F, Alam, I. | | Deposit date: | 2014-12-01 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Identification of Non-nucleoside Human Ribonucleotide Reductase Modulators.
J.Med.Chem., 58, 2015
|
|
5OLK
 
 | | Crystal structure of the ATP-cone-containing NrdB from Leeuwenhoekiella blandensis | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MAGNESIUM ION, ... | | Authors: | Hasan, M, Grinberg, A.R, Sjoberg, B.M, Logan, D.T. | | Deposit date: | 2017-07-28 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Novel ATP-cone-driven allosteric regulation of ribonucleotide reductase via the radical-generating subunit.
Elife, 7, 2018
|
|
7AGJ
 
 | | Ribonucleotide Reductase R1 protein from Aquifex aeolicus | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Rehling, D, Scaletti, E.R, Stenmark, P. | | Deposit date: | 2020-09-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Biochemical Investigation of Class I Ribonucleotide Reductase from the Hyperthermophile Aquifex aeolicus.
Biochemistry, 61, 2022
|
|
6R1R
 
 | | RIBONUCLEOTIDE REDUCTASE E441D MUTANT R1 PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | RIBONUCLEOTIDE REDUCTASE R1 PROTEIN, RIBONUCLEOTIDE REDUCTASE R2 PROTEIN | | Authors: | Eriksson, M, Eklund, H. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase.
J.Biol.Chem., 272, 1997
|
|
2R1R
 
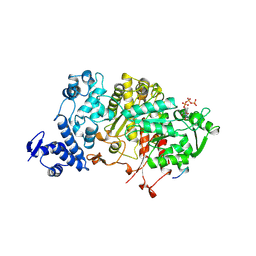 | |
7MDI
 
 | | Structure of the Neisseria gonorrhoeae ribonucleotide reductase in the inactive state | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CYTIDINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Levitz, T.S, Drennan, C.L, Brignole, E.J. | | Deposit date: | 2021-04-05 | | Release date: | 2022-01-05 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Effects of chameleon dispense-to-plunge speed on particle concentration, complex formation, and final resolution: A case study using the Neisseria gonorrhoeae ribonucleotide reductase inactive complex.
J.Struct.Biol., 214, 2021
|
|
4R1R
 
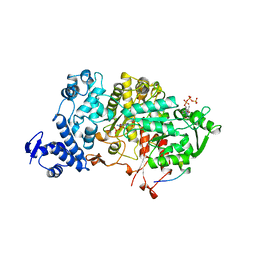 | |
1ZYZ
 
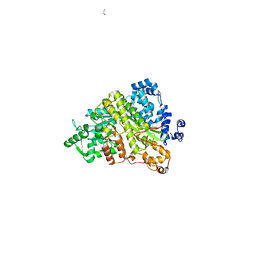 | | Structures of Yeast Ribonucloetide Reductase I | | Descriptor: | GLYCINE, Ribonucleoside-diphosphate reductase large chain 1 | | Authors: | Xu, H, Faber, C, Uchiki, T, Fairman, J.W, Racca, J, Dealwis, C. | | Deposit date: | 2005-06-13 | | Release date: | 2006-03-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of eukaryotic ribonucleotide reductase I provide insights into dNTP regulation
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5D1Y
 
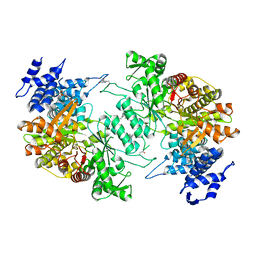 | | Low resolution crystal structure of human ribonucleotide reductase alpha6 hexamer in complex with dATP | | Descriptor: | Ribonucleoside-diphosphate reductase large subunit | | Authors: | Ando, N, Li, H, Brignole, E.J, Thompson, S, McLaughlin, M.I, Page, J, Asturias, F, Stubbe, J, Drennan, C.L. | | Deposit date: | 2015-08-04 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (9.005 Å) | | Cite: | Allosteric Inhibition of Human Ribonucleotide Reductase by dATP Entails the Stabilization of a Hexamer.
Biochemistry, 55, 2016
|
|
5R1R
 
 | | RIBONUCLEOTIDE REDUCTASE E441A MUTANT R1 PROTEIN FROM ESCHERICHIA COLI | | Descriptor: | RIBONUCLEOTIDE REDUCTASE R1 PROTEIN, RIBONUCLEOTIDE REDUCTASE R2 PROTEIN | | Authors: | Eriksson, M, Eklund, H. | | Deposit date: | 1997-09-17 | | Release date: | 1998-03-18 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new mechanism-based radical intermediate in a mutant R1 protein affecting the catalytically essential Glu441 in Escherichia coli ribonucleotide reductase.
J.Biol.Chem., 272, 1997
|
|
1R1R
 
 | |
1RLR
 
 | |
