6QFM
 
 | | Structure of human Mcl-1 in complex with PUMA BH3 peptide | | Descriptor: | Bcl-2-binding component 3, CHLORIDE ION, Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QZ5
 
 | | Structure of Mcl-1 in complex with compound 8a | | Descriptor: | (2~{R})-2-[[6-ethyl-5-(1~{H}-indol-4-yl)thieno[2,3-d]pyrimidin-4-yl]amino]-3-phenyl-propanoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
4WMX
 
 | | The structure of MBP-MCL1 bound to ligand 6 at 2.0A | | Descriptor: | 4-ethenyl-2-[(phenylsulfonyl)amino]benzoic acid, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
5II4
 
 | | Crystal structure of red abalone VERL repeat 1 with linker at 2.0 A resolution | | Descriptor: | Maltose-binding periplasmic protein,Vitelline envelope sperm lysin receptor, TRIETHYLENE GLYCOL, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
6YBK
 
 | | Structure of MBP-Mcl-1 in complex with compound 4d | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-(pyrazin-2-ylmethoxy)phenyl]propanoic acid, CHLORIDE ION, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Dokurno, P, Surgenor, A.E, Murray, J.B. | | Deposit date: | 2020-03-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of S64315, a Potent and Selective Mcl-1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
5TTD
 
 | | Minor pilin FctB from S. pyogenes with engineered intramolecular isopeptide bond | | Descriptor: | FORMIC ACID, Maltose-binding periplasmic protein,Pilin isopeptide linkage domain protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Young, P.G, Kwon, H, Squire, C.J, Baker, E.N. | | Deposit date: | 2016-11-02 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering a Lys-Asn isopeptide bond into an immunoglobulin-like protein domain enhances its stability.
Sci Rep, 7, 2017
|
|
4YS9
 
 | |
5GPP
 
 | | Crystal structure of zebrafish ASC PYD domain | | Descriptor: | ACETATE ION, Maltose-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, ... | | Authors: | Jin, T, Li, Y. | | Deposit date: | 2016-08-04 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Functional and structural characterization of zebrafish ASC.
FEBS J., 285, 2018
|
|
2PQK
 
 | | X-ray crystal structure of human Mcl-1 in complex with Bim BH3 | | Descriptor: | Bim BH3 peptide, Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Bare, E, Grant, R.A, Keating, A.E. | | Deposit date: | 2007-05-02 | | Release date: | 2007-06-19 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes.
Protein Sci., 19, 2010
|
|
3OSR
 
 | | Maltose-bound maltose sensor engineered by insertion of circularly permuted green fluorescent protein into E. coli maltose binding protein at position 311 | | Descriptor: | Maltose-binding periplasmic protein,Green fluorescent protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Echevarria, I.M, Marvin, J.S, Looger, L.L, Schreiter, E.R. | | Deposit date: | 2010-09-09 | | Release date: | 2011-10-26 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A genetically encoded, high-signal-to-noise maltose sensor.
Proteins, 79, 2011
|
|
5BMY
 
 | |
3D7V
 
 | | Crystal structure of Mcl-1 in complex with an Mcl-1 selective BH3 ligand | | Descriptor: | Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Lee, E.F, Czabotar, P.E, Colman, P.M, Fairlie, W.D. | | Deposit date: | 2008-05-22 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A novel BH3 ligand that selectively targets Mcl-1 reveals that apoptosis can proceed without Mcl-1 degradation.
J.Cell Biol., 180, 2008
|
|
5JON
 
 | | Crystal structure of the unliganded form of HCN2 CNBD | | Descriptor: | Maltose-binding periplasmic protein,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2, NITRATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Klenchin, V.A, Chanda, B. | | Deposit date: | 2016-05-02 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Structure and dynamics underlying elementary ligand binding events in human pacemaking channels.
Elife, 5, 2016
|
|
4WVG
 
 | |
5C6H
 
 | | Mcl-1 complexed with Mule | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, Mule BH3 peptide from E3 ubiquitin-protein ligase HUWE1 | | Authors: | Song, T, Wang, Z, Ji, F, Chai, G, Liu, Y, Li, X, Li, Z, Fan, Y, Zhang, Z. | | Deposit date: | 2015-06-23 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Mcl-1 complexed with Mule at 2.05 Angstroms resolution
To Be Published
|
|
6D1U
 
 | | Crystal structure of the human CLR:RAMP1 extracellular domain heterodimer in complex with adrenomedullin 2/intermedin | | Descriptor: | ADM2, Maltose-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, SODIUM ION, ... | | Authors: | Pioszak, A, Roehrkasse, A. | | Deposit date: | 2018-04-12 | | Release date: | 2018-09-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-function analyses reveal a triple beta-turn receptor-bound conformation of adrenomedullin 2/intermedin and enable peptide antagonist design.
J. Biol. Chem., 293, 2018
|
|
3VD8
 
 | | Crystal structure of human AIM2 PYD domain with MBP fusion | | Descriptor: | 1,2-ETHANEDIOL, Maltose-binding periplasmic protein, Interferon-inducible protein AIM2, ... | | Authors: | Jin, T.C, Perry, A, Smith, P, Xiao, T.S. | | Deposit date: | 2012-01-04 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0685 Å) | | Cite: | Structure of the Absent in Melanoma 2 (AIM2) Pyrin Domain Provides Insights into the Mechanisms of AIM2 Autoinhibition and Inflammasome Assembly.
J.Biol.Chem., 288, 2013
|
|
6U65
 
 | | Mcl-1 bound to compound 19 | | Descriptor: | 1,2-ETHANEDIOL, 2-[({4-[(4-fluorophenyl)methyl]piperazin-1-yl}sulfonyl)amino]-5-[(2-phenylethyl)sulfanyl]benzoic acid, ACETATE ION, ... | | Authors: | Stuckey, J.A. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
6UAB
 
 | |
6QYO
 
 | | Structure of MBP-Mcl-1 in complex with compound 18a | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-ethyl-thieno[2,3-d]pyrimidin-4-yl]oxy-3-phenyl-propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Szlavik, Z, Ondi, L, Csekei, M, Paczal, A, Szabo, Z.B, Radics, G, Murray, J, Davidson, J, Chen, I, Davis, B, Hubbard, R.E, Pedder, C, Surgenor, A.E, Smith, J, Robertson, A, LeToumelin-Braizat, G, Cauquil, N, Zarka, M, Demarles, D, Perron-Sierra, F, Geneste, O, Kotschy, A. | | Deposit date: | 2019-03-09 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Discovery of a Selective Mcl-1 Inhibitor with Cellular Activity.
J.Med.Chem., 62, 2019
|
|
4WVH
 
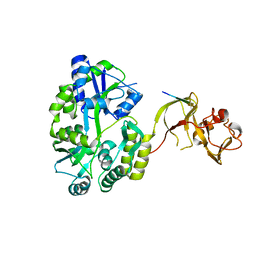 | | Crystal structure of the Type-I signal peptidase from Staphylococcus aureus (SpsB) in complex with a substrate peptide (pep1). | | Descriptor: | Maltose-binding periplasmic protein,Signal peptidase IB, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, substrate peptide (pep1) | | Authors: | Young, P.G, Ting, Y.T, Baker, E.N. | | Deposit date: | 2014-11-05 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peptide binding to a bacterial signal peptidase visualized by peptide tethering and carrier-driven crystallization.
IUCrJ, 3, 2016
|
|
5FIO
 
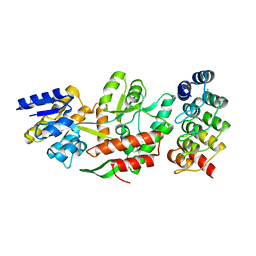 | | DARPins as a new tool for experimental phasing in protein crystallography | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN, MERCURY (II) ION, NI3C DARPIN MUTANT5 HG-SITE N1 | | Authors: | Batyuk, A, Honegger, A, Andres, F, Briand, C, Gruetter, M, Plueckthun, A. | | Deposit date: | 2015-09-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Darpins as a New Tool for Experimental Phasing in Protein Crystallography
To be Published
|
|
6YBL
 
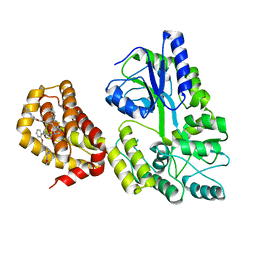 | | Structure of MBP-Mcl-1 in complex with compound 9m | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-[[2-(2-methoxyphenyl)pyrimidin-4-yl]methoxy]phenyl]propanoic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Dokurno, P, Surgenor, A.E, Murray, J.B. | | Deposit date: | 2020-03-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of S64315, a Potent and Selective Mcl-1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
6YBG
 
 | | Structure of Mcl-1 in complex with compound 2g | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(3-chlorophenyl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-(2-methoxyphenyl)propanoic acid, CHLORIDE ION, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Dokurno, P, Surgenor, A.E, Murray, J.B. | | Deposit date: | 2020-03-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of S64315, a Potent and Selective Mcl-1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
5T03
 
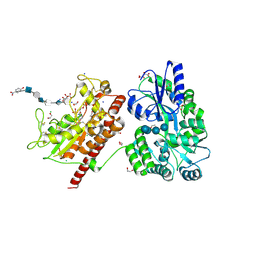 | | Crystal structure of heparan sulfate 6-O-sulfotransferase with bound PAP and glucuronic acid containing hexasaccharide substrate | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Pedersen, L.C, Moon, A.F, Krahn, J.M, Liu, J. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
