4M9E
 
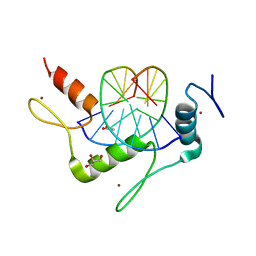 | | Structure of Klf4 zinc finger DNA binding domain in complex with methylated DNA | | 分子名称: | ACETATE ION, DNA (5'-D(*GP*AP*GP*GP*(5CM)P*GP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*(5CM)P*GP*CP*CP*TP*C)-3'), ... | | 著者 | Liu, Y, Olanrewaju, Y.O, Blumenthal, R.M, Zhang, X, Cheng, X. | | 登録日 | 2013-08-14 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.851 Å) | | 主引用文献 | Structural basis for Klf4 recognition of methylated DNA.
Nucleic Acids Res., 42, 2014
|
|
2WBS
 
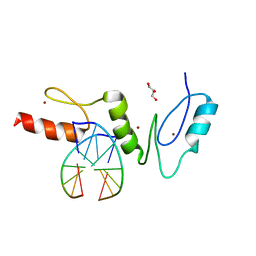 | | Crystal structure of the zinc finger domain of Klf4 bound to its target DNA | | 分子名称: | 5'-D(*GP*AP*GP*GP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*CP*CP*TP*CP)-3', GLYCEROL, ... | | 著者 | Zocher, G, Schuetz, A, Carstanjen, D, Heinemann, U. | | 登録日 | 2009-03-03 | | 公開日 | 2010-04-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The Structure of the Klf4 DNA-Binding Domain Links to Self-Renewal and Macrophage Differentiation.
Cell.Mol.Life Sci., 68, 2011
|
|
2WBU
 
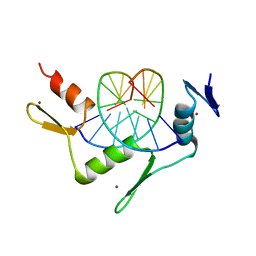 | | CRYSTAL STRUCTURE OF THE ZINC FINGER DOMAIN OF KLF4 BOUND TO ITS TARGET DNA | | 分子名称: | 5'-D(*DGP*DAP*DGP*DGP*DCP*DGP*DTP* DGP*DGP*DC)-3', 5'-D(*DGP*DCP*DCP*DAP*DCP*DGP*DCP* DCP*DTP*DC)-3', KRUEPPEL-LIKE FACTOR 4, ... | | 著者 | Schuetz, A, Zocher, G, Carstanjen, D, Heinemann, U. | | 登録日 | 2009-03-05 | | 公開日 | 2010-04-07 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Structure of the Klf4 DNA-Binding Domain Links to Self-Renewal and Macrophage Differentiation.
Cell.Mol.Life Sci., 68, 2011
|
|
6VTX
 
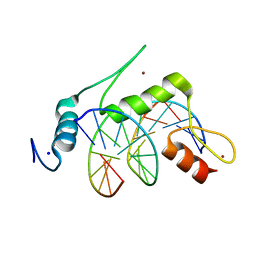 | | Crystal structure of human KLF4 zinc finger DNA binding domain in complex with NANOG DNA | | 分子名称: | DNA (5'-D(*AP*GP*GP*GP*GP*GP*TP*GP*TP*GP*CP*C)-3'), DNA (5'-D(*GP*GP*CP*AP*CP*AP*CP*CP*CP*CP*CP*T)-3'), Krueppel-like factor 4, ... | | 著者 | Sharma, R, Sharma, S, Choi, K.J, Ferreon, A.C.M, Ferreon, J.C, Sankaran, B, MacKenzie, K.R, Kim, C. | | 登録日 | 2020-02-13 | | 公開日 | 2021-09-01 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Liquid condensation of reprogramming factor KLF4 with DNA provides a mechanism for chromatin organization.
Nat Commun, 12, 2021
|
|
5KE8
 
 | |
5KE9
 
 | |
5KEA
 
 | |
5KE7
 
 | |
5KE6
 
 | |
5KEB
 
 | |
6UZK
 
 | |
6OQ4
 
 | |
6OQ3
 
 | |
3L1P
 
 | | POU protein:DNA complex | | 分子名称: | DNA (5'-D(*AP*TP*CP*CP*AP*TP*TP*TP*GP*CP*CP*TP*TP*TP*CP*AP*AP*AP*TP*GP*TP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*AP*CP*AP*TP*TP*TP*GP*AP*AP*AP*GP*GP*CP*AP*AP*AP*TP*GP*GP*A)-3'), POU domain, ... | | 著者 | Vahokoski, J, Groves, M.R, Pogenberg, V, Wilmanns, M. | | 登録日 | 2009-12-14 | | 公開日 | 2010-12-15 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | A unique Oct4 interface is crucial for reprogramming to pluripotency
Nat.Cell Biol., 15, 2013
|
|
3X1U
 
 | |
3X1V
 
 | |
3X1T
 
 | |
3X1S
 
 | | Crystal structure of the nucleosome core particle | | 分子名称: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | 著者 | Sivaraman, P, Kumarevel, T.S. | | 登録日 | 2014-11-27 | | 公開日 | 2015-09-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.805 Å) | | 主引用文献 | Structural and functional analyses of nucleosome complexes with mouse histone variants TH2a and TH2b, involved in reprogramming
Biochem.Biophys.Res.Commun., 464, 2015
|
|
6OQZ
 
 | | Crystal structure of Glucose Isomerase from Non-merohedrally twinned crystals | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, MAGNESIUM ION, MANGANESE (II) ION, ... | | 著者 | Sevvana, M, Ruf, M, Uson, I, Sheldrick, G.M, Herbst-Irmer, R. | | 登録日 | 2019-04-29 | | 公開日 | 2019-12-11 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Non-merohedral twinning: from minerals to proteins.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
