3LDU
 
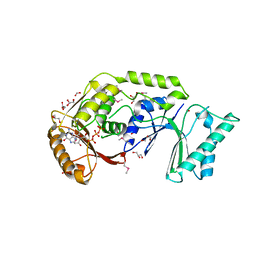 | | The crystal structure of a possible methylase from Clostridium difficile 630. | | 分子名称: | FORMIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Tan, K, Wu, R, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-01-13 | | 公開日 | 2010-01-26 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The crystal structure of a possible methylase from Clostridium difficile 630.
To be Published
|
|
3LED
 
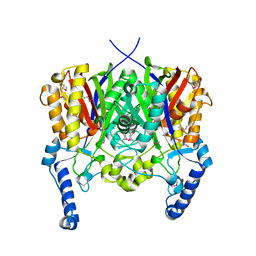 | | Crystal structure of 3-oxoacyl-(acyl carrier protein) synthase III from Rhodopseudomonas palustris CGA009 | | 分子名称: | 3-oxoacyl-acyl carrier protein synthase III, FORMIC ACID | | 著者 | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2010-01-14 | | 公開日 | 2010-01-26 | | 最終更新日 | 2017-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal structure of 3-oxoacyl-(acyl carrier protein) synthase III from Rhodopseudomonas palustris CGA009
To be Published
|
|
5H69
 
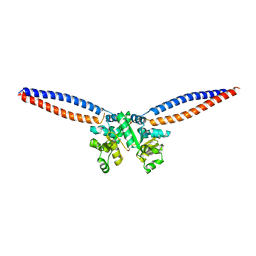 | |
5HCJ
 
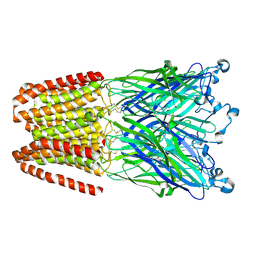 | | Cationic Ligand-Gated Ion Channel | | 分子名称: | CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, Proton-gated ion channel, ... | | 著者 | Shahsavar, A, Sauguet, L, Delarue, M. | | 登録日 | 2016-01-04 | | 公開日 | 2016-04-13 | | 最終更新日 | 2016-04-20 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Sites of Anesthetic Inhibitory Action on a Cationic Ligand-Gated Ion Channel.
Structure, 24, 2016
|
|
5HD3
 
 | |
6UH9
 
 | |
3L7J
 
 | |
5H6K
 
 | | DNA targeting ADP-ribosyltransferase Pierisin-1 | | 分子名称: | 1,2-ETHANEDIOL, Pierisin-1 | | 著者 | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | 登録日 | 2016-11-14 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
3LFJ
 
 | |
5H6V
 
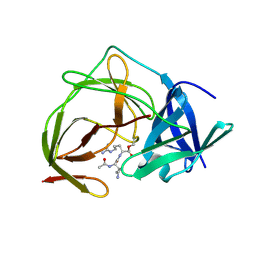 | |
5HDA
 
 | |
3L8J
 
 | | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity | | 分子名称: | Programmed cell death protein 10 | | 著者 | Li, X, Zhang, R, Zhang, H, He, Y, Ji, W, Min, W, Boggon, T.J. | | 登録日 | 2009-12-31 | | 公開日 | 2010-05-19 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity.
J.Biol.Chem., 285, 2010
|
|
5HDE
 
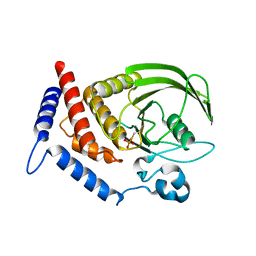 | |
3LHK
 
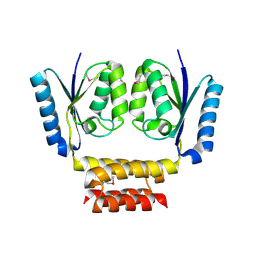 | |
5H72
 
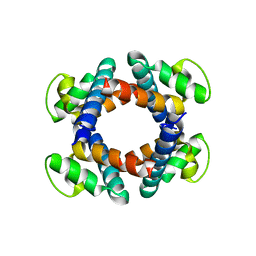 | | Structure of the periplasmic domain of FliP | | 分子名称: | Flagellar biosynthetic protein FliP | | 著者 | Fukumura, T, Kawaguchi, T, Saijo-Hamano, Y, Namba, K, Minamino, T, Imada, K. | | 登録日 | 2016-11-16 | | 公開日 | 2017-08-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex
PLoS Biol., 15, 2017
|
|
5HDL
 
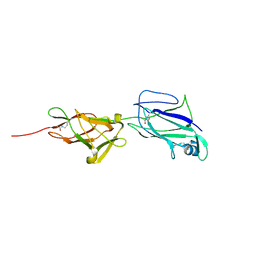 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG - E269A mutant | | 分子名称: | Cell surface protein SpaA | | 著者 | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | 登録日 | 2016-01-05 | | 公開日 | 2016-07-20 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
3LI1
 
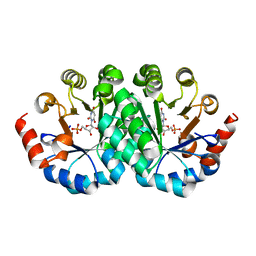 | | Crystal structure of the mutant I218A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | 分子名称: | 1-(5'-PHOSPHO-BETA-D-RIBOFURANOSYL)BARBITURIC ACID, Orotidine 5'-phosphate decarboxylase | | 著者 | Fedorov, A.A, Fedorov, E.V, Wood, B.M, Gerlt, J.A, Almo, S.C. | | 登録日 | 2010-01-23 | | 公開日 | 2010-06-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Conformational changes in orotidine 5'-monophosphate decarboxylase: "remote" residues that stabilize the active conformation.
Biochemistry, 49, 2010
|
|
5H79
 
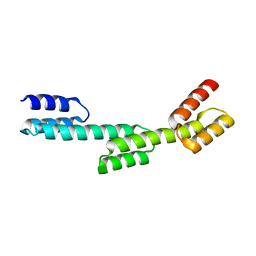 | | Crystal structure of a repeat protein with three Protein A repeat module | | 分子名称: | Immunoglobulin G-binding protein A | | 著者 | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | 登録日 | 2016-11-17 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
3LIG
 
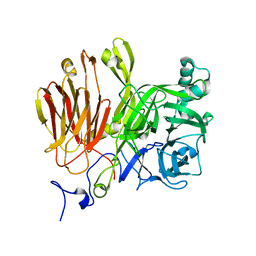 | |
5H7F
 
 | |
5HDT
 
 | |
3LIX
 
 | | crystal structure of htlv protease complexed with the inhibitor KNI-10729 | | 分子名称: | N-{(1S,2S)-1-benzyl-3-[(4R)-5,5-dimethyl-4-{[(1R)-1,2,2-trimethylpropyl]carbamoyl}-1,3-thiazolidin-3-yl]-2-hydroxy-3-oxopropyl}-3-methyl-N~2~-{(2S)-2-[(morpholin-4-ylacetyl)amino]-2-phenylacetyl}-L-valinamide, Protease, ZINC ION | | 著者 | Satoh, T, Li, M, Nguyen, J, Kiso, Y, Wlodawer, A, Gustchina, A. | | 登録日 | 2010-01-25 | | 公開日 | 2010-07-14 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal structures of inhibitor complexes of human T-cell leukemia virus (HTLV-1) protease.
J.Mol.Biol., 401, 2010
|
|
5HEW
 
 | |
5H88
 
 | | Crystal structure of mRojoA mutant - T16V -P63F - W143A - L163V | | 分子名称: | mRojoA fluorescent protein | | 著者 | Pandelieva, A.T, Tremblay, V, Sarvan, S, Chica, R.A, Couture, J.-F. | | 登録日 | 2015-12-23 | | 公開日 | 2016-01-27 | | 最終更新日 | 2016-03-02 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Brighter Red Fluorescent Proteins by Rational Design of Triple-Decker Motif.
Acs Chem.Biol., 11, 2016
|
|
3LH0
 
 | |
