3OIA
 
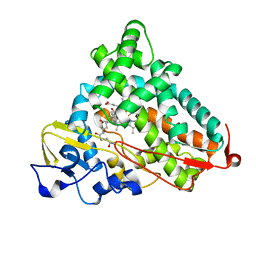 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C8GluEtg-Bio | | 分子名称: | Camphor 5-monooxygenase, N-{15-oxo-19-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-4,7,10-trioxa-14-azanonadec-1-yl}-N'-(8-{[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-ylcarbonyl]amino}octyl)pentanediamide, POTASSIUM ION, ... | | 著者 | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | 登録日 | 2010-08-18 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C8GluEtg-Bio
To be Published
|
|
1G0L
 
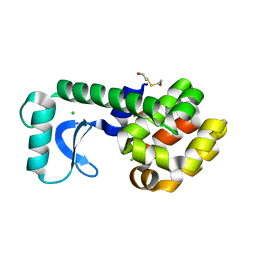 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT T152V | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | 著者 | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | 登録日 | 2000-10-06 | | 公開日 | 2001-05-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
5JWS
 
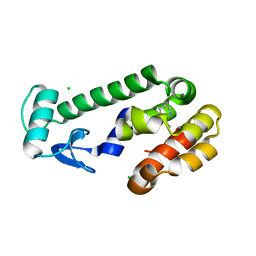 | | T4 Lysozyme L99A with 1-Hydro-2-ethyl-1,2-azaborine Bound | | 分子名称: | 2-ethyl-1,2-dihydro-1,2-azaborinine, CHLORIDE ION, Endolysin | | 著者 | Lee, H, Fischer, M, Shoichet, B.K, Liu, S.-Y. | | 登録日 | 2016-05-12 | | 公開日 | 2016-09-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Hydrogen Bonding of 1,2-Azaborines in the Binding Cavity of T4 Lysozyme Mutants: Structures and Thermodynamics.
J.Am.Chem.Soc., 138, 2016
|
|
4WJ9
 
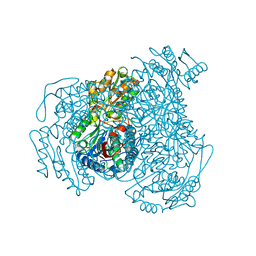 | | Structure of Human apo ALDH1A1 | | 分子名称: | CHLORIDE ION, Retinal dehydrogenase 1, YTTERBIUM (III) ION | | 著者 | Morgan, C.A, Hurley, T.D. | | 登録日 | 2014-09-29 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Development of a high-throughput in vitro assay to identify selective inhibitors for human ALDH1A1.
Chem.Biol.Interact., 234, 2015
|
|
3ORF
 
 | | Crystal Structure of Dihydropteridine Reductase from Dictyostelium discoideum | | 分子名称: | Dihydropteridine reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Chen, C, Zhuang, N.N, Seo, K.H, Park, Y.S, Lee, K.H. | | 登録日 | 2010-09-07 | | 公開日 | 2011-07-27 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Structural insights into the dual substrate specificities of mammalian and Dictyostelium dihydropteridine reductases toward two stereoisomers of quinonoid dihydrobiopterin
Febs Lett., 585, 2011
|
|
1G07
 
 | | CRYSTAL STRUCTURE OF T4 LYSOZYME MUTANT V149C | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, PROTEIN (LYSOZYME) | | 著者 | Xu, J, Baase, W.A, Quillin, M.L, Matthews, B.W. | | 登録日 | 2000-10-05 | | 公開日 | 2001-05-23 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural and thermodynamic analysis of the binding of solvent at internal sites in T4 lysozyme.
Protein Sci., 10, 2001
|
|
4WKG
 
 | |
3OL5
 
 | | Crystal Structure of Cytochrome P450cam crystallized with a tethered substrate analog 3OH-AdaC1-C8-Dans | | 分子名称: | (1R,3S,5R,7S)-N-[8-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)octyl]-3-hydroxytricyclo[3.3.1.1~3,7~]decane-1-carboxamide, Camphor 5-monooxygenase, POTASSIUM ION, ... | | 著者 | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | 登録日 | 2010-08-25 | | 公開日 | 2010-11-17 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structure of Cytochrome P450cam crystallized with a tethered substrate analog 3OH-AdaC1-C8-Dans
To be Published
|
|
4WMZ
 
 | | S. cerevisiae CYP51 complexed with fluconazole in the active site | | 分子名称: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Sagatova, A, Keniya, M.V, Wilson, R, Wilbanks, S.M, Monk, B.C, Tyndall, J.D.A. | | 登録日 | 2014-10-09 | | 公開日 | 2015-07-01 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Structural Insights into Binding of the Antifungal Drug Fluconazole to Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase.
Antimicrob.Agents Chemother., 59, 2015
|
|
5KF6
 
 | |
3OLI
 
 | | Structures of human pancreatic alpha-amylase in complex with acarviostatin IV03 | | 分子名称: | (1S,2S,3R,6R)-6-amino-4-(hydroxymethyl)cyclohex-4-ene-1,2,3-triol, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | 著者 | Qin, X, Ren, L. | | 登録日 | 2010-08-26 | | 公開日 | 2011-04-13 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structures of human pancreatic alpha-amylase in complex with acarviostatins: Implications for drug design against type II diabetes.
J.Struct.Biol., 174, 2011
|
|
4WOJ
 
 | | Aspartate Semialdehyde Dehydrogenase from Francisella tularensis | | 分子名称: | Aspartate semialdehyde dehydrogenase, SODIUM ION, SULFATE ION | | 著者 | Mank, N.J, Arnette, A.K, Klapper, V.G, Chruszcz, M. | | 登録日 | 2014-10-15 | | 公開日 | 2015-10-21 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Structure of aspartate b-semialdehyde dehydrogenase from Francisella tularensis
Acta Crystallogr.,Sect.F, 74, 2018
|
|
5JP0
 
 | | Bacteroides ovatus Xyloglucan PUL GH3B with bound glucose | | 分子名称: | Beta-glucosidase BoGH3B, MAGNESIUM ION, beta-D-glucopyranose | | 著者 | Hemsworth, G.R, Thompson, A.J, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | 登録日 | 2016-05-03 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
1UMD
 
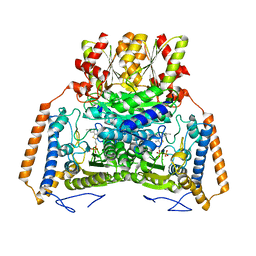 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 with 4-methyl-2-oxopentanoate as an intermediate | | 分子名称: | 2-OXO-4-METHYLPENTANOIC ACID, 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, ... | | 著者 | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-09-25 | | 公開日 | 2004-03-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
7EXK
 
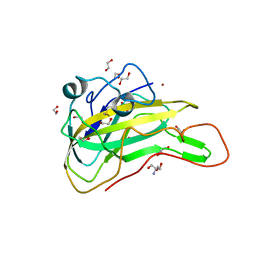 | | An AA9 LPMO of Ceriporiopsis subvermispora | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Nguyen, H, Kondo, K, Nagata, T, Katahira, M, Mikami, B. | | 登録日 | 2021-05-27 | | 公開日 | 2022-05-04 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.14 Å) | | 主引用文献 | Functional and Structural Characterizations of Lytic Polysaccharide Monooxygenase, Which Cooperates Synergistically with Cellulases, from Ceriporiopsis subvermispora.
Acs Sustain Chem Eng, 10, 2022
|
|
4LRS
 
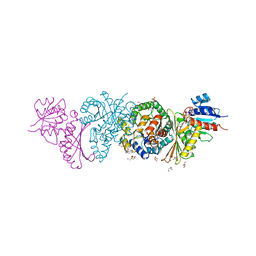 | | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site | | 分子名称: | 4-hydroxy-2-oxovalerate aldolase, Acetaldehyde dehydrogenase, CHLORIDE ION, ... | | 著者 | Fischer, B, Branlant, G, Talfournier, F, Gruez, A. | | 登録日 | 2013-07-20 | | 公開日 | 2013-09-04 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Crystal and solution structures of the bifunctional enzyme (Aldolase/Aldehyde dehydrogenase) from Thermomonospora curvata, reveal a cofactor-binding domain motion during NAD+ and CoA accommodation whithin the shared cofactor-binding site
To be Published
|
|
5JTA
 
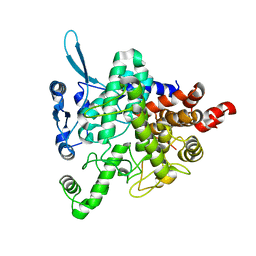 | |
3OOQ
 
 | | CRYSTAL STRUCTURE OF amidohydrolase from Thermotoga maritima MSB8 | | 分子名称: | GLYCEROL, amidohydrolase | | 著者 | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-08-31 | | 公開日 | 2010-09-15 | | 最終更新日 | 2021-02-10 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF amidohydrolase from Thermotoga maritima MSB8
To be Published
|
|
7EUN
 
 | |
7EUQ
 
 | |
4WKT
 
 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | 分子名称: | 1-BUTANE BORONIC ACID, Acyl-homoserine lactone acylase PvdQ, GLYCEROL | | 著者 | Wu, R, Clevenger, K.D, Fast, W, Liu, D. | | 登録日 | 2014-10-03 | | 公開日 | 2014-11-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.782 Å) | | 主引用文献 | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
3ORV
 
 | |
4LMR
 
 | | Crystal structure of L-lactate dehydrogenase from Bacillus cereus ATCC 14579, NYSGRC Target 029452 | | 分子名称: | L-lactate dehydrogenase 1 | | 著者 | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2013-07-10 | | 公開日 | 2013-07-31 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | Crystal structure of L-lactate dehydrogenase from Bacillus cereus ATCC 14579, NYSGRC Target 029452
To be Published
|
|
4WVY
 
 | |
3OVR
 
 | | Crystal Structure of hRPE and D-Xylulose 5-Phosphate Complex | | 分子名称: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, 5-O-phosphono-D-xylulose, FE (II) ION, ... | | 著者 | Liang, W.G, Ouyang, S.Y, Shaw, N, Joachimiak, A, Zhang, R.G, Liu, Z.J. | | 登録日 | 2010-09-17 | | 公開日 | 2011-03-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.948 Å) | | 主引用文献 | Conversion of D-ribulose 5-phosphate to D-xylulose 5-phosphate: new insights from structural and biochemical studies on human RPE
Faseb J., 25, 2011
|
|
