4QVI
 
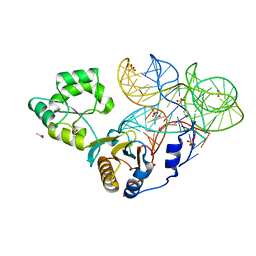 | | Crystal structure of mutant ribosomal protein M218L TthL1 in complex with 80nt 23S RNA from Thermus thermophilus | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 50S ribosomal protein L1, ACETATE ION, ... | | 著者 | Gabdulkhakov, A.G, Nevskaya, N.A, NIkonov, S.V. | | 登録日 | 2014-07-15 | | 公開日 | 2015-02-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4R5P
 
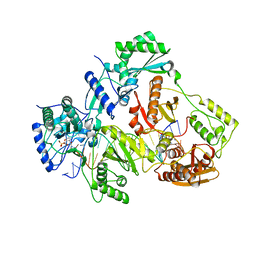 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and a nucleoside triphosphate mimic alpha-carboxy nucleoside phosphonate inhibitor | | 分子名称: | 5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*G)-3', 5'-D(*TP*GP*GP*AP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3', HIV-1 reverse transcriptase, ... | | 著者 | Das, K, Martinez, S.E, Arnold, E. | | 登録日 | 2014-08-21 | | 公開日 | 2015-03-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.894 Å) | | 主引用文献 | Alpha-carboxy nucleoside phosphonates as universal nucleoside triphosphate mimics.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RES
 
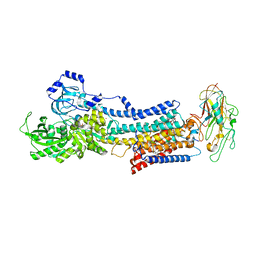 | | Crystal structure of the Na,K-ATPase E2P-bufalin complex with bound potassium | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Laursen, M, Yatime, L, Gregersen, J.L, Nissen, P, Fedosova, N.U. | | 登録日 | 2014-09-23 | | 公開日 | 2015-01-28 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.408 Å) | | 主引用文献 | Structures and characterization of digoxin- and bufalin-bound Na+,K+-ATPase compared with the ouabain-bound complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RET
 
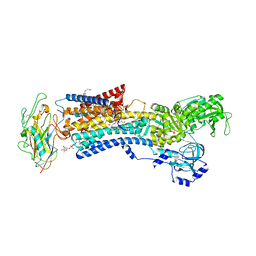 | | Crystal structure of the Na,K-ATPase E2P-digoxin complex with bound magnesium | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | 著者 | Gregersen, J.L, Laursen, M, Yatime, L, Nissen, P, Fedosova, N.U. | | 登録日 | 2014-09-23 | | 公開日 | 2015-01-28 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | Structures and characterization of digoxin- and bufalin-bound Na+,K+-ATPase compared with the ouabain-bound complex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4RK5
 
 | | Crystal structure of LacI family transcriptional regulator from Lactobacillus casei, Target EFI-512911, with bound sucrose | | 分子名称: | ACETATE ION, SODIUM ION, Transcriptional regulator, ... | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-10-11 | | 公開日 | 2014-11-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Crystal Structure of LacI Transcriptional Regulator Lsei_2103 from Lactobacillus casei, Target EFI-512911
To be Published
|
|
4RK7
 
 | | Crystal structure of LacI family transcriptional regulator CCPA from Weissella paramesenteroides, Target EFI-512926, with bound sucrose | | 分子名称: | Glucose-resistance amylase regulator, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | 著者 | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | 登録日 | 2014-10-11 | | 公開日 | 2014-10-29 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of transcriptional regulator CCPA from Weissella paramesenteroides, Target EFI-512926
To be Published
|
|
4RYK
 
 | | Crystal structure of a putative transcriptional regulator from Listeria monocytogenes EGD-e | | 分子名称: | DI(HYDROXYETHYL)ETHER, L(+)-TARTARIC ACID, Lmo0325 protein, ... | | 著者 | Filippova, E.V, Wawrzak, Z, Minasov, G, Kiryukhina, O, Jedrzejczak, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2014-12-15 | | 公開日 | 2015-01-07 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.09 Å) | | 主引用文献 | Crystal structure of a putative transcriptional regulator from Listeria monocytogenes EGD-e
To be Published
|
|
4TS0
 
 | | Crystal structure of the Spinach RNA aptamer in complex with DFHBI, barium ions | | 分子名称: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, BARIUM ION, POTASSIUM ION, ... | | 著者 | Warner, K.D, Chen, M.C, Song, W, Strack, R.L, Thorn, A, Jaffrey, S.R, Ferre-D'Amare, A.R. | | 登録日 | 2014-06-18 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structural basis for activity of highly efficient RNA mimics of green fluorescent protein.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4TS2
 
 | | Crystal structure of the Spinach RNA aptamer in complex with DFHBI, magnesium ions | | 分子名称: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, MAGNESIUM ION, POTASSIUM ION, ... | | 著者 | Warner, K.D, Chen, M.C, Song, W, Strack, R.L, Thorn, A, Jaffrey, S.R, Ferre-D'Amare, A.R. | | 登録日 | 2014-06-18 | | 公開日 | 2014-07-23 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.884 Å) | | 主引用文献 | Structural basis for activity of highly efficient RNA mimics of green fluorescent protein.
Nat.Struct.Mol.Biol., 21, 2014
|
|
4UQG
 
 | | A new bio-isosteric base pair based on reversible bonding | | 分子名称: | 5'-D(*AP*GP*GP*GP*A SAYP*GP*GP*TP*CP)-3', 5'-D(*GP*AP*CP*C T0TP*TP*CP*CP*CP*TP)-3', DNA POLYMERASE, ... | | 著者 | Tomas-Gamasa, M, Serdjukov, S, Su, M, Mueller, M, Carell, T. | | 登録日 | 2014-06-23 | | 公開日 | 2014-12-03 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | "Post-it" type connected DNA created with a reversible covalent cross-link.
Angew. Chem. Int. Ed. Engl., 54, 2015
|
|
4YFU
 
 | |
4YWP
 
 | |
4ZE9
 
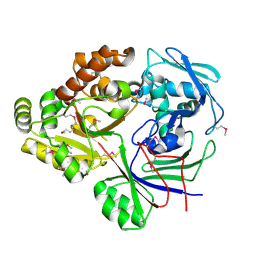 | |
4ZNO
 
 | | Crystal structure of Dln1 complexed with sucrose | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Natterin-like protein, ... | | 著者 | Jia, N, Jiang, Y.L, Cheng, W, Wang, H.W, Zhou, C.Z, Chen, Y. | | 登録日 | 2015-05-05 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein.
Embo Rep., 17, 2016
|
|
4ZXW
 
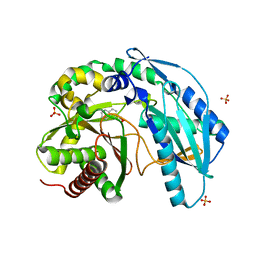 | | Crystal structure of SgcC5 protein from Streptomyces globisporus (complex with (R)-(-)-1-(2-naphthyl)-1,2-ethanediol and sucrose) | | 分子名称: | (1R)-1-(naphthalen-2-yl)ethane-1,2-diol, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, C-domain type II peptide synthetase, ... | | 著者 | Michalska, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | 登録日 | 2015-05-20 | | 公開日 | 2015-06-17 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.187 Å) | | 主引用文献 | Crystal structure of SgcC5 protein from Streptomyces globisporus
To Be Published
|
|
5D2D
 
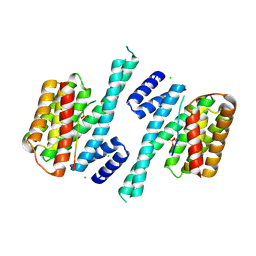 | | Crystal structure of human 14-3-3 zeta in complex with CFTR R-domain peptide pS753-pS768 | | 分子名称: | 14-3-3 protein zeta/delta, CHLORIDE ION, Cystic fibrosis transmembrane conductance regulator, ... | | 著者 | Stevers, L.M, Leysen, S.F.R, Ottmann, C. | | 登録日 | 2015-08-05 | | 公開日 | 2016-03-16 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Characterization and small-molecule stabilization of the multisite tandem binding between 14-3-3 and the R domain of CFTR.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5D3G
 
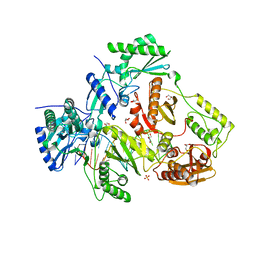 | | Structure of HIV-1 Reverse Transcriptase Bound to a Novel 38-mer Hairpin Template-Primer DNA Aptamer | | 分子名称: | DNA aptamer (38-MER), GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | 著者 | Miller, M.T, Tuske, S, Das, K, Arnold, E. | | 登録日 | 2015-08-06 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of HIV-1 reverse transcriptase bound to a novel 38-mer hairpin template-primer DNA aptamer.
Protein Sci., 25, 2016
|
|
5DK3
 
 | | Crystal Structure of Pembrolizumab, a full length IgG4 antibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain, ... | | 著者 | Scapin, G, Prosise, W, Reichert, P. | | 登録日 | 2015-09-02 | | 公開日 | 2015-11-18 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Structure of full-length human anti-PD1 therapeutic IgG4 antibody pembrolizumab.
Nat.Struct.Mol.Biol., 22, 2015
|
|
5E58
 
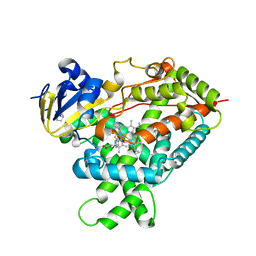 | | Crystal Structure Of Cytochrome P450 2B35 from Desert Woodrat Neotoma Lepida in complex with 4-(4-chlorophenyl)imidazole | | 分子名称: | 4-(4-CHLOROPHENYL)IMIDAZOLE, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 family 2 subfamily B, ... | | 著者 | Shah, M.B, Stout, C.D, Halpert, J.R. | | 登録日 | 2015-10-08 | | 公開日 | 2016-02-10 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structure-Function Analysis of Mammalian CYP2B Enzymes Using 7-Substituted Coumarin Derivatives as Probes: Utility of Crystal Structures and Molecular Modeling in Understanding Xenobiotic Metabolism.
Mol.Pharmacol., 89, 2016
|
|
5FIX
 
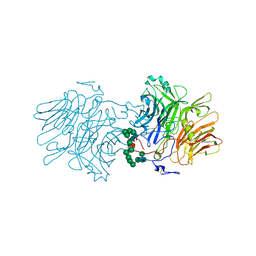 | |
5GOP
 
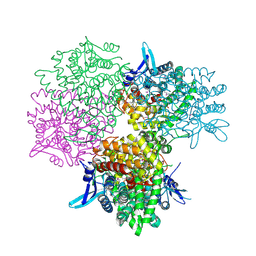 | | Crystal structure of alkaline invertase InvA from Anabaena sp. PCC 7120 complexed with sucrose | | 分子名称: | Alkaline Invertase, beta-D-fructofuranose, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | 著者 | Xie, J, Cai, K, Hu, H.X, Jiang, Y.L, Yang, F, Hu, P.F, Chen, Y, Zhou, C.Z. | | 登録日 | 2016-07-28 | | 公開日 | 2016-11-02 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structural Analysis of the Catalytic Mechanism and Substrate Specificity of Anabaena Alkaline Invertase InvA Reveals a Novel Glucosidase
J. Biol. Chem., 291, 2016
|
|
5HLF
 
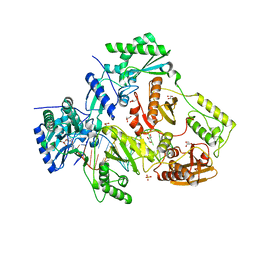 | |
5HP1
 
 | |
5HRO
 
 | |
5I3U
 
 | |
