8E6G
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 10 | | 分子名称: | (2R,3R,4R,5S)-1-(6-{[(5M)-3-cyclopropyl-5-(pyridazin-3-yl)phenyl]amino}hexyl)-2-(hydroxymethyl)piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Karade, S.S, Mariuzza, R.A. | | 登録日 | 2022-08-22 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EBM
 
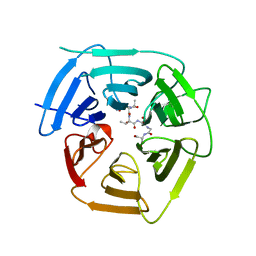 | |
6LXI
 
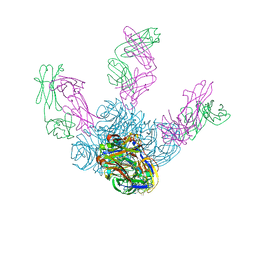 | | Crystal structure of Z2B3 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1) | | 分子名称: | CALCIUM ION, Heavy chain of Z2B3 Fab, Light chain of Z2B3 Fab, ... | | 著者 | Jiang, H, Peng, W, Qi, J, Chai, Y, Song, H, Shi, Y, Gao, G.F, Wu, Y. | | 登録日 | 2020-02-11 | | 公開日 | 2020-12-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure-Based Modification of an Anti-neuraminidase Human Antibody Restores Protection Efficacy against the Drifted Influenza Virus.
Mbio, 11, 2020
|
|
4ZCW
 
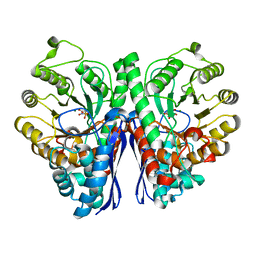 | | Structure of Human Enolase 2 in complex with SF2312 | | 分子名称: | Gamma-enolase, MAGNESIUM ION, [(3S,5S)-1,5-dihydroxy-2-oxopyrrolidin-3-yl]phosphonic acid | | 著者 | Leonard, P.G, Maxwell, D, Czako, B, Muller, F.L. | | 登録日 | 2015-04-16 | | 公開日 | 2016-07-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.992 Å) | | 主引用文献 | SF2312 is a natural phosphonate inhibitor of enolase.
Nat.Chem.Biol., 12, 2016
|
|
4ZDR
 
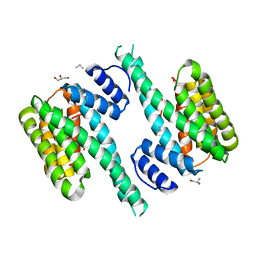 | | Crystal structure of 14-3-3[zeta]-LKB1 fusion protein | | 分子名称: | 14-3-3 protein zeta/delta,GGSGGS linker,Serine/threonine-protein kinase STK11, GLYCEROL, PROPANE, ... | | 著者 | Ding, S, Shi, Z.B. | | 登録日 | 2015-04-18 | | 公開日 | 2015-09-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.899 Å) | | 主引用文献 | Structure of the 14-3-3 zeta-LKB1 fusion protein provides insight into a novel ligand-binding mode of 14-3-3.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8EBN
 
 | |
8B3T
 
 | | Hen Egg White Lysozyme 4s in situ crystallization | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Henkel, A, Galchenkova, M, Yefanov, O, Hakanpaeae, J, Chapman, H.N, Oberthuer, D. | | 登録日 | 2022-09-16 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | JINXED: just in time crystallization for easy structure determination of biological macromolecules.
Iucrj, 10, 2023
|
|
6BUQ
 
 | |
7MLO
 
 | | Crystal structure of ricin A chain in complex with 5-mesitylthiophene-2-carboxylic acid | | 分子名称: | 1,2-ETHANEDIOL, 5-(2,4,6-trimethylphenyl)thiophene-2-carboxylic acid, Ricin, ... | | 著者 | Harijan, R.K, Li, X.P, Cao, B, Augeri, D, Bonanno, J.B, Almo, S.C, Tumer, N.E, Schramm, V.L. | | 登録日 | 2021-04-28 | | 公開日 | 2022-02-09 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Synthesis and Structural Characterization of Ricin Inhibitors Targeting Ribosome Binding Using Fragment-Based Methods and Structure-Based Design.
J.Med.Chem., 64, 2021
|
|
3BE1
 
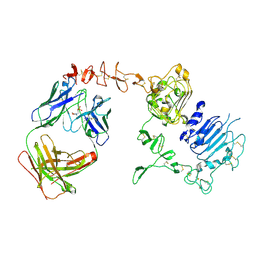 | | Dual specific bH1 Fab in complex with the extracellular domain of HER2/ErbB-2 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab Fragment-Heavy Chain, ... | | 著者 | Bostrom, J.M, Wiesmann, C, Appleton, B.A. | | 登録日 | 2007-11-15 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Variants of the antibody herceptin that interact with HER2 and VEGF at the antigen binding site
Science, 323, 2009
|
|
8B3U
 
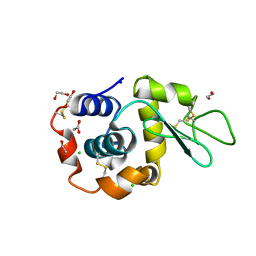 | | Hen Egg White Lysozyme 6s in situ crystallization | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Henkel, A, Galchenkova, M, Yefanov, O, Hakanpaeae, J, Chapman, H.N, Oberthuer, D. | | 登録日 | 2022-09-16 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | JINXED: just in time crystallization for easy structure determination of biological macromolecules.
Iucrj, 10, 2023
|
|
6BS6
 
 | |
6IBQ
 
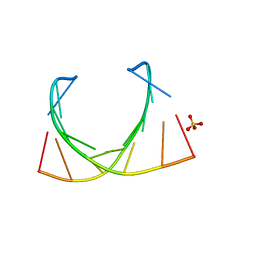 | | Structure of a nonameric RNA duplex at room temperature in ChipX microfluidic device | | 分子名称: | DNA/RNA (5'-R(*CP*GP*UP*GP*AP*UP*CP*G)-D(P*C)-3'), SULFATE ION | | 著者 | de Wijn, R, Olieric, V, Lorber, B, Sauter, C. | | 登録日 | 2018-11-30 | | 公開日 | 2019-05-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | A simple and versatile microfluidic device for efficient biomacromolecule crystallization and structural analysis by serial crystallography.
Iucrj, 6, 2019
|
|
8DKA
 
 | |
6M4M
 
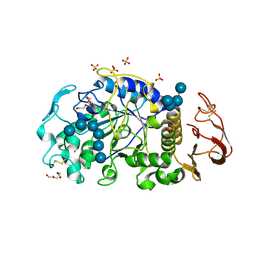 | | X-ray crystal structure of the E249Q mutan of alpha-amylase I and maltohexaose complex from Eisenia fetida | | 分子名称: | Alpha-amylase, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Hirano, Y, Tsukamoto, K, Ariki, S, Naka, Y, Ueda, M, Tamada, T. | | 登録日 | 2020-03-07 | | 公開日 | 2020-09-16 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | X-ray crystallographic structural studies of alpha-amylase I from Eisenia fetida.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8KGY
 
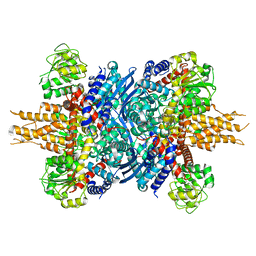 | | Human glutamate dehydrogenase I | | 分子名称: | Glutamate dehydrogenase 1, mitochondrial | | 著者 | Su, M.-Y. | | 登録日 | 2023-08-20 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | ELECTRON MICROSCOPY (2.59 Å) | | 主引用文献 | Cryo-EM structure of the KLHL22 E3 ligase bound to an oligomeric metabolic enzyme.
Structure, 31, 2023
|
|
8B3V
 
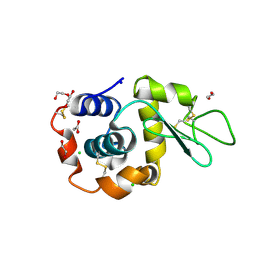 | | Hen Egg White Lysozyme 8s in situ crystallization | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | 著者 | Henkel, A, Galchenkova, M, Yefanov, O, Hakanpaeae, J, Chapman, H.N, Oberthuer, D. | | 登録日 | 2022-09-16 | | 公開日 | 2022-11-30 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | JINXED: just in time crystallization for easy structure determination of biological macromolecules.
Iucrj, 10, 2023
|
|
6BUV
 
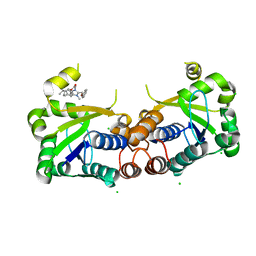 | | Structure of Mycobacterium tuberculosis NadD in complex with inhibitor [(1~{R},2~{R},5~{S})-5-methyl-2-propan-2-yl-cyclohexyl] 2-[3-methyl-2-(phenoxymethyl)benzimidazol-1-yl]ethanoate | | 分子名称: | 1-methyl-3-(2-{[(1R,2R,5S)-5-methyl-2-(propan-2-yl)cyclohexyl]oxy}-2-oxoethyl)-2-(phenoxymethyl)-1H-1,3-benzimidazol-3-ium, CHLORIDE ION, SODIUM ION, ... | | 著者 | Rodionova, I.A, Reed, R.W, Sorci, L, Osterman, A.L, Korotkov, K.V. | | 登録日 | 2017-12-11 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | Novel Antimycobacterial Compounds Suppress NAD Biogenesis by Targeting a Unique Pocket of NaMN Adenylyltransferase.
Acs Chem.Biol., 14, 2019
|
|
6VA8
 
 | |
7MJC
 
 | |
7MJD
 
 | | Crystal Structure Analysis of ALDH1B1 | | 分子名称: | 8-(2-methoxyphenyl)-10-(4-phenylphenyl)-1$l^{4},8-diazabicyclo[5.3.0]deca-1(7),9-diene, Aldehyde dehydrogenase X, mitochondrial, ... | | 著者 | Fernandez, D, Chen, J.K. | | 登録日 | 2021-04-20 | | 公開日 | 2022-06-08 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Inhibitors targeted to aldehyde dehydrogenase
Nat.Chem.Biol., 2022
|
|
8E4K
 
 | | Co-crystal structure of Chaetomium glucosidase with compound 7 | | 分子名称: | (2R,3R,4R,5S)-2-(hydroxymethyl)-1-[6-({(4M)-4-[2-(morpholin-4-yl)pyrimidin-4-yl]-2-nitrophenyl}amino)hexyl]piperidine-3,4,5-triol, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Karade, S.S, Mariuzza, R.A. | | 登録日 | 2022-08-18 | | 公開日 | 2023-02-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure-Based Design of Potent Iminosugar Inhibitors of Endoplasmic Reticulum alpha-Glucosidase I with Anti-SARS-CoV-2 Activity.
J.Med.Chem., 66, 2023
|
|
8EBL
 
 | |
6BW3
 
 | | Crystal structure of RBBP4 in complex with PRDM3 N-terminal peptide | | 分子名称: | Histone-binding protein RBBP4, MDS1 and EVI1 complex locus protein MDS1, UNKNOWN ATOM OR ION | | 著者 | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | 登録日 | 2017-12-14 | | 公開日 | 2017-12-27 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
1F6O
 
 | | CRYSTAL STRUCTURE OF THE HUMAN AAG DNA REPAIR GLYCOSYLASE COMPLEXED WITH DNA | | 分子名称: | 3-METHYL-ADENINE DNA GLYCOSYLASE, DNA (5'-D(*GP*AP*CP*AP*TP*GP*(YRR)P*TP*TP*GP*CP*CP*T)-3'), DNA (5'-D(*GP*GP*CP*AP*AP*TP*CP*AP*TP*GP*TP*CP*A)-3'), ... | | 著者 | Lau, A.Y, Wyatt, M.D, Glassner, B.J, Samson, L.D, Ellenberger, T. | | 登録日 | 2000-06-22 | | 公開日 | 2000-12-11 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
