8C6A
 
 | | Light SFX structure of D.m(6-4)photolyase at 1ps time delay | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | 登録日 | 2023-01-11 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6B
 
 | | Light SFX structure of D.m(6-4)photolyase at 20ps time delay | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | 登録日 | 2023-01-11 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6F
 
 | | Light SFX structure of D.m(6-4)photolyase at 400fs time delay | | 分子名称: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | 著者 | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | 登録日 | 2023-01-11 | | 公開日 | 2023-11-01 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C2F
 
 | |
7UE1
 
 | | HIV-1 Integrase Catalytic Core Domain Mutant (KGD) in Complex with Inhibitor GRL-142 | | 分子名称: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, Integrase, SULFATE ION | | 著者 | Aoki, M, Aoki-Ogata, H, Bulut, H, Hayashi, H, Davis, D, Hasegawa, K, Yarchoan, R, Ghosh, A.K, Pau, A.K, Mitsuya, H. | | 登録日 | 2022-03-21 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-04-24 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | GRL-142 binds to and impairs HIV-1 integrase nuclear localization signal and potently suppresses highly INSTI-resistant HIV-1 variants.
Sci Adv, 9, 2023
|
|
5AGQ
 
 | | Solution structure of the TAM domain of human TIP5 BAZ2A involved in epigenetic regulation of rRNA genes | | 分子名称: | BROMODOMAIN ADJACENT TO ZINC FINGER DOMAIN PROTEIN 2A | | 著者 | Anosova, I, Melnik, S, Tripsianes, K, Kateb, F, Grummt, I, Sattler, M. | | 登録日 | 2015-02-03 | | 公開日 | 2015-05-06 | | 最終更新日 | 2024-06-19 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Novel RNA Binding Surface of the Tam Domain of Tip5/Baz2A Mediates Epigenetic Regulation of Rrna Genes.
Nucleic Acids Res., 43, 2015
|
|
5A7R
 
 | | Human poly(ADP-ribose) glycohydrolase in complex with synthetic dimeric ADP-ribose | | 分子名称: | BETA-MERCAPTOETHANOL, GLYCEROL, POLY(ADP-RIBOSE) GLYCOHYDROLASE, ... | | 著者 | Lambrecht, M.J, Brichacek, M, Barkauskaite, E, Ariza, A, Ahel, I, Hergenrother, P.J. | | 登録日 | 2015-07-09 | | 公開日 | 2015-07-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Synthesis of Dimeric Adp-Ribose and its Structure with Human Poly(Adp-Ribose) Glycohydrolase.
J.Am.Chem.Soc., 137, 2015
|
|
8DGF
 
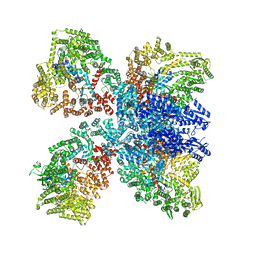 | | Avs4 bound to phage PhiV-1 portal | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding protein Avs4, MAGNESIUM ION, ... | | 著者 | Wilkinson, M.E, Gao, L, Strecker, J, Makarova, K.S, Macrae, R.K, Koonin, E.V, Zhang, F. | | 登録日 | 2022-06-23 | | 公開日 | 2022-08-03 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Prokaryotic innate immunity through pattern recognition of conserved viral proteins.
Science, 377, 2022
|
|
4ZXQ
 
 | |
8CRG
 
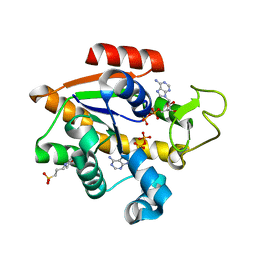 | | E. coli adenylate kinase in complex with two ADP molecules as a result of enzymatic AP4A hydrolysis | | 分子名称: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase | | 著者 | Oelker, M, Tischlik, S, Wolf-Watz, M, Sauer-Eriksson, A.E. | | 登録日 | 2023-03-08 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Insights into Enzymatic Catalysis from Binding and Hydrolysis of Diadenosine Tetraphosphate by E. coli Adenylate Kinase.
Biochemistry, 62, 2023
|
|
5BON
 
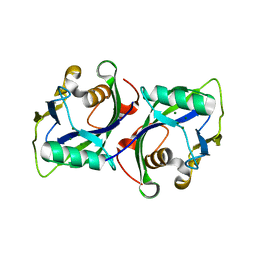 | | Crystal structure of human Nudt15 (MTH2) | | 分子名称: | MAGNESIUM ION, Probable 8-oxo-dGTP diphosphatase NUDT15 | | 著者 | Carter, M, Jemth, A.-S, Helleday, T, Stenmark, P. | | 登録日 | 2015-05-27 | | 公開日 | 2015-08-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.799 Å) | | 主引用文献 | Crystal structure, biochemical and cellular activities demonstrate separate functions of MTH1 and MTH2.
Nat Commun, 6, 2015
|
|
8E1I
 
 | | Asp1 kinase in complex with ATP Mg 5-IP7 | | 分子名称: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, ADENOSINE-5'-TRIPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, ... | | 著者 | Goldgur, Y, Shuman, S, Benjamin, B. | | 登録日 | 2022-08-10 | | 公開日 | 2022-11-30 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
8E5T
 
 | | Yeast co-transcriptional Noc1-Noc2 RNP assembly checkpoint intermediate | | 分子名称: | 25S ribosomal RNA, 5.8S ribosomal RNA, 60S ribosomal protein L13-A, ... | | 著者 | Sanghai, Z.A, Piwowarczyk, R, Vanden Broeck, A, Klinge, S. | | 登録日 | 2022-08-22 | | 公開日 | 2023-04-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | A co-transcriptional ribosome assembly checkpoint controls nascent large ribosomal subunit maturation.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8EB7
 
 | |
1Y98
 
 | | Structure of the BRCT repeats of BRCA1 bound to a CtIP phosphopeptide. | | 分子名称: | Breast cancer type 1 susceptibility protein, COBALT (II) ION, CtIP PHOSPHORYLATED PEPTIDE, ... | | 著者 | Varma, A.K, Brown, R.S, Birrane, G, Ladias, J.A.A. | | 登録日 | 2004-12-14 | | 公開日 | 2005-08-30 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural Basis for Cell Cycle Checkpoint Control by the BRCA1-CtIP Complex.
Biochemistry, 44, 2005
|
|
6I8T
 
 | |
1WFQ
 
 | | Solution structure of the first cold-shock domain of the human KIAA0885 protein (UNR protein) | | 分子名称: | UNR protein | | 著者 | Goroncy, A.K, Kigawa, T, Koshiba, S, Tomizawa, T, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2004-05-26 | | 公開日 | 2004-11-26 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
6J0D
 
 | | Crystal structure of OsSUF4 | | 分子名称: | ZINC ION, transcription factor | | 著者 | Wang, B, Luo, Q. | | 登録日 | 2018-12-24 | | 公開日 | 2019-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The transcription factor OsSUF4 interacts with SDG725 in promoting H3K36me3 establishment.
Nat Commun, 10, 2019
|
|
6J0W
 
 | | Crystal Structure of Yeast Rtt107 and Nse6 | | 分子名称: | Peptide from DNA repair protein KRE29, Regulator of Ty1 transposition protein 107 | | 著者 | Wan, B, Wu, J, Lei, M. | | 登録日 | 2018-12-27 | | 公開日 | 2019-08-14 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Molecular Basis for Control of Diverse Genome Stability Factors by the Multi-BRCT Scaffold Rtt107.
Mol.Cell, 75, 2019
|
|
5CNV
 
 | | Crystal structure of the dATP inhibited E. coli class Ia ribonucleotide reductase complex bound to GDP and TTP at 3.20 Angstroms resolution | | 分子名称: | 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Chen, P.Y.-T, Zimanyi, C.M, Funk, M.A, Drennan, C.L. | | 登録日 | 2015-07-18 | | 公開日 | 2016-01-20 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Molecular basis for allosteric specificity regulation in class Ia ribonucleotide reductase from Escherichia coli.
Elife, 5, 2016
|
|
2RS7
 
 | | Solution structure of the second dsRBD from RNA helicase A | | 分子名称: | ATP-dependent RNA helicase A | | 著者 | Nagata, T, Muto, Y, Tsuda, K, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2011-11-29 | | 公開日 | 2012-03-14 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures of the double-stranded RNA-binding domains from RNA helicase A
Proteins, 80, 2012
|
|
2RS6
 
 | | Solution structure of the N-terminal dsRBD from RNA helicase A | | 分子名称: | ATP-dependent RNA helicase A | | 著者 | Nagata, T, Muto, Y, Tsuda, K, Inoue, M, Kigawa, T, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2011-11-29 | | 公開日 | 2012-03-14 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures of the double-stranded RNA-binding domains from RNA helicase A
Proteins, 80, 2012
|
|
6H4C
 
 | | A polyamorous repressor: deciphering the evolutionary strategy used by the phage-inducible chromosomal islands to spread in nature. | | 分子名称: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, NICKEL (II) ION, ... | | 著者 | Ciges-Tomas, J.R, Alite, C, Bowring, J.Z, Donderis, J, Penades, J.R, Marina, A. | | 登録日 | 2018-07-20 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.52 Å) | | 主引用文献 | The structure of a polygamous repressor reveals how phage-inducible chromosomal islands spread in nature.
Nat Commun, 10, 2019
|
|
6JCF
 
 | | Cryogenic structure of HIV-1 Integrase catalytic core domain by synchrotron | | 分子名称: | CACODYLATE ION, Integrase | | 著者 | Park, J.H, Han, J, Kim, T.H, Yun, J.H, Lee, W. | | 登録日 | 2019-01-28 | | 公開日 | 2019-07-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.153 Å) | | 主引用文献 | Non-Cryogenic Structure and Dynamics of HIV-1 Integrase Catalytic Core Domain by X-ray Free-Electron Lasers.
Int J Mol Sci, 20, 2019
|
|
1YDX
 
 | | Crystal structure of Type-I restriction-modification system S subunit from M. genitalium | | 分子名称: | CHLORIDE ION, type I restriction enzyme specificity protein MG438 | | 著者 | Machado, B, Quijada, O, Pinol, J, Fita, I, Querol, E, Carpena, X. | | 登録日 | 2004-12-27 | | 公開日 | 2005-08-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structure of a Putative Type I Restriction-Modification S Subunit from Mycoplasma genitalium
J.Mol.Biol., 351, 2005
|
|
