1OCD
 
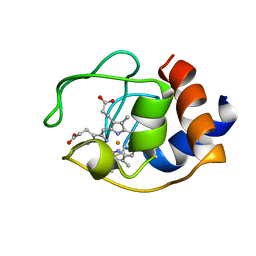 | | CYTOCHROME C (OXIDIZED) FROM EQUUS CABALLUS, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | CYTOCHROME C, HEME C | | 著者 | Qi, P.X, Beckman, R.A, Wand, A.J. | | 登録日 | 1996-07-03 | | 公開日 | 1997-06-16 | | 最終更新日 | 2018-03-14 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of horse heart ferricytochrome c and detection of redox-related structural changes by high-resolution 1H NMR.
Biochemistry, 35, 1996
|
|
2PCF
 
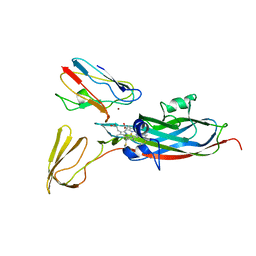 | | THE COMPLEX OF CYTOCHROME F AND PLASTOCYANIN DETERMINED WITH PARAMAGNETIC NMR. BASED ON THE STRUCTURES OF CYTOCHROME F AND PLASTOCYANIN, 10 STRUCTURES | | 分子名称: | COPPER (II) ION, CYTOCHROME F, HEME C, ... | | 著者 | Ubbink, M, Ejdeback, M, Karlsson, B.G, Bendall, D.S. | | 登録日 | 1997-12-22 | | 公開日 | 1998-04-08 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The structure of the complex of plastocyanin and cytochrome f, determined by paramagnetic NMR and restrained rigid-body molecular dynamics.
Structure, 6, 1998
|
|
1P5P
 
 | |
1P5O
 
 | |
1P5M
 
 | |
1P5N
 
 | |
1BC4
 
 | | THE SOLUTION STRUCTURE OF A CYTOTOXIC RIBONUCLEASE FROM THE OOCYTES OF RANA CATESBEIANA (BULLFROG), NMR, 15 STRUCTURES | | 分子名称: | RIBONUCLEASE | | 著者 | Chang, C.-F, Chen, C, Chen, Y.-C, Hom, K, Huang, R.-F, Huang, T. | | 登録日 | 1998-05-05 | | 公開日 | 1998-10-14 | | 最終更新日 | 2019-12-25 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of a cytotoxic ribonuclease from the oocytes of Rana catesbeiana (bullfrog).
J.Mol.Biol., 283, 1998
|
|
1KOD
 
 | | RNA APTAMER COMPLEXED WITH CITRULLINE, NMR | | 分子名称: | CITRULLINE, RNA (5'-R(P*AP*CP*GP*GP*UP*UP*AP*GP*GP*UP*CP*GP*CP*U)-3'), RNA (5'-R(P*AP*GP*AP*AP*GP*GP*AP*GP*UP*GP*U)-3') | | 著者 | Yang, Y.S, Kochoyan, M, Burgstaller, P, Westhof, E, Famulok, M. | | 登録日 | 1996-03-28 | | 公開日 | 1996-11-08 | | 最終更新日 | 2023-11-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structural basis of ligand discrimination by two related RNA aptamers resolved by NMR spectroscopy.
Science, 272, 1996
|
|
1OT4
 
 | | Solution structure of Cu(II)-CopC from Pseudomonas syringae | | 分子名称: | COPPER (II) ION, Copper resistance protein C | | 著者 | Arnesano, F, Banci, L, Bertini, I, Felli, I.C, Luchinat, C, Thompsett, A.R, Structural Proteomics in Europe (SPINE) | | 登録日 | 2003-03-21 | | 公開日 | 2003-07-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | A Strategy for the NMR Characterization of Type II Copper(II) Proteins:
the Case of the Copper Trafficking Protein CopC from Pseudomonas Syringae.
J.Am.Chem.Soc., 125, 2003
|
|
1BM6
 
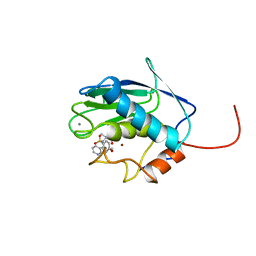 | | SOLUTION STRUCTURE OF THE CATALYTIC DOMAIN OF HUMAN STROMELYSIN-1 COMPLEXED TO A POTENT NON-PEPTIDIC INHIBITOR, NMR, 20 STRUCTURES | | 分子名称: | 1-METHYLOXY-4-SULFONE-BENZENE, 3-METHYLPYRIDINE, CALCIUM ION, ... | | 著者 | Li, Y, Zhang, X, Melton, R, Ganu, V, Gonnella, N.C. | | 登録日 | 1998-07-29 | | 公開日 | 1999-07-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the catalytic domain of human stromelysin-1 complexed to a potent, nonpeptidic inhibitor.
Biochemistry, 37, 1998
|
|
1RIP
 
 | |
1SE9
 
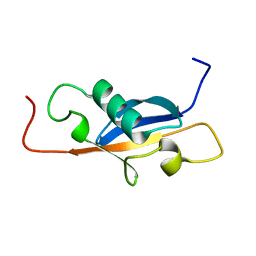 | |
1PGA
 
 | |
1PGB
 
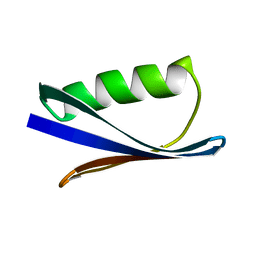 | |
1E09
 
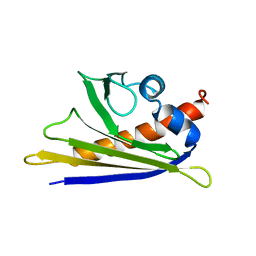 | | Solution Structure of the Major Cherry Allergen Pru av 1 | | 分子名称: | PRU AV 1 | | 著者 | Neudecker, P, Nerkamp, J, Schweimer, K, Sticht, H, Boehm, M, Scheurer, S, Vieths, S, Roesch, P. | | 登録日 | 2000-03-15 | | 公開日 | 2001-03-15 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Allergic Cross-Reactivity Made Visible: The Solution Structure of the Major Cherry Allergen Pru Av 1
J.Biol.Chem., 276, 2001
|
|
3GBQ
 
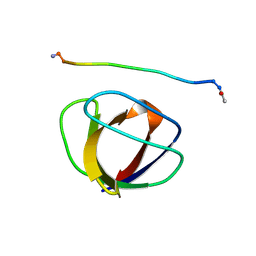 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | GRB2, SOS-1 | | 著者 | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | 登録日 | 1996-12-23 | | 公開日 | 1997-09-04 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
1KUN
 
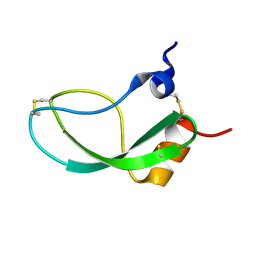 | | SOLUTION STRUCTURE OF THE HUMAN ALPHA3-CHAIN TYPE VI COLLAGEN C-TERMINAL KUNITZ DOMAIN, NMR, 20 STRUCTURES | | 分子名称: | ALPHA3-CHAIN TYPE VI COLLAGEN | | 著者 | Sorensen, M.D, Bjorn, S, Norris, K, Olsen, O, Petersen, L, James, T.L, Led, J.J. | | 登録日 | 1997-03-04 | | 公開日 | 1997-11-12 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and backbone dynamics of the human alpha3-chain type VI collagen C-terminal Kunitz domain,.
Biochemistry, 36, 1997
|
|
8ULM
 
 | |
1NEH
 
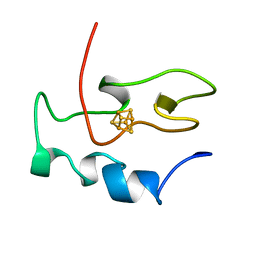 | | HIGH POTENTIAL IRON-SULFUR PROTEIN | | 分子名称: | HIGH POTENTIAL IRON SULFUR PROTEIN, IRON/SULFUR CLUSTER | | 著者 | Bertini, I, Dikiy, A, Kastrau, D.H.W, Luchinat, C, Sompornpisut, P. | | 登録日 | 1995-12-14 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Three-dimensional solution structure of the oxidized high potential iron-sulfur protein from Chromatium vinosum through NMR. Comparative analysis with the solution structure of the reduced species.
Biochemistry, 34, 1995
|
|
2RR3
 
 | | Solution structure of the complex between human VAP-A MSP domain and human OSBP FFAT motif | | 分子名称: | Oxysterol-binding protein 1, Vesicle-associated membrane protein-associated protein A | | 著者 | Furuita, K, Jee, J, Fukada, H, Mishima, M, Kojima, C. | | 登録日 | 2010-03-09 | | 公開日 | 2010-03-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Electrostatic interaction between oxysterol-binding protein and VAMP-associated protein A revealed by NMR and mutagenesis studies
J.Biol.Chem., 285, 2010
|
|
2RN1
 
 | | Liquid crystal solution structure of the kissing complex formed by the apical loop of the HIV TAR RNA and a high affinity RNA aptamer optimized by SELEX | | 分子名称: | RNA (5'-R(P*GP*AP*GP*CP*CP*CP*UP*GP*GP*GP*AP*GP*GP*CP*UP*C)-3'), RNA (5'-R(P*GP*CP*UP*GP*GP*UP*CP*CP*CP*AP*GP*AP*CP*AP*GP*C)-3') | | 著者 | Van Melckebeke, H, Devany, M, Di Primo, C, Beaurain, F, Toulme, J, Bryce, D.L, Boisbouvier, J. | | 登録日 | 2007-12-05 | | 公開日 | 2008-09-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Liquid-crystal NMR structure of HIV TAR RNA bound to its SELEX RNA aptamer reveals the origins of the high stability of the complex
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3AIT
 
 | |
1I25
 
 | |
5UPW
 
 | |
6GN4
 
 | | tc-DNA/tc-DNA duplex | | 分子名称: | Tc-DNA (5'-D(*(TCJ)P*(TTK)P*(TCJ)P*(TCS)P*(TCS)P*(TCJ)P*(TTK)P*(TTK)P*(TCY)P*(TCJ))-3'), Tc-DNA (5'-D(*(TCS)P*(TTK)P*(TCY)P*(TCY)P*(TCS)P*(TCJ)P*(TCJ)P*(TCS)P*(TCY)P*(TCS))-3') | | 著者 | Istrate, A, Johannsen, S, Istrate, A, Sigel, R.K.O, Leumann, C. | | 登録日 | 2018-05-29 | | 公開日 | 2018-06-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of tricyclo-DNA containing duplexes: insight into enhanced thermal stability and nuclease resistance.
Nucleic Acids Res., 47, 2019
|
|
