4ASN
 
 | |
2MWO
 
 | |
8UX1
 
 | |
5TUS
 
 | | Potent competitive inhibition of human ribonucleotide reductase by a novel non-nucleoside small molecule | | 分子名称: | 2-hydroxy-N'-[(Z)-(2-hydroxynaphthalen-1-yl)methylidene]benzohydrazide, MAGNESIUM ION, Ribonucleoside-diphosphate reductase large subunit, ... | | 著者 | Mohammed, F.A, Alam, I, Dealwis, C.G. | | 登録日 | 2016-11-07 | | 公開日 | 2017-08-02 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.66 Å) | | 主引用文献 | Potent competitive inhibition of human ribonucleotide reductase by a nonnucleoside small molecule.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2IO5
 
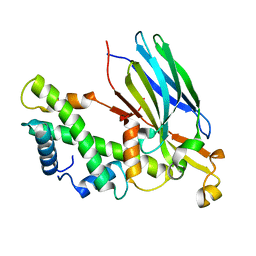 | | Crystal structure of the CIA- histone H3-H4 complex | | 分子名称: | ASF1A protein, Histone H3.1, Histone H4 | | 著者 | Natsume, R, Akai, Y, Horikoshi, M, Senda, T. | | 登録日 | 2006-10-10 | | 公開日 | 2007-02-27 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4.
Nature, 446, 2007
|
|
2MWP
 
 | |
8A5C
 
 | |
8A5F
 
 | |
8PK1
 
 | | Cas1-Cas2 CRISPR integrase bound to prespacer DNA, Streptococcus thermophilus DGCC 7710 CRISPR3 system | | 分子名称: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, Chains: G,J, ... | | 著者 | Sasnauskas, G, Gaizauskaite, U, Tamulaitiene, G. | | 登録日 | 2023-06-23 | | 公開日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Structural basis for spacer acquisition in a type II-A CRISPR-Cas system
To Be Published
|
|
5FSL
 
 | | MTH1 substrate recognition: Complex with a methylaminopurinone | | 分子名称: | 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, 9-METHYL-2-(METHYLAMINO)-1H-PURIN-6-ONE, SULFATE ION | | 著者 | Nissink, J.W.M, Bista, M, Breed, J, Carter, N, Embrey, K, Read, J, Phillips, C, Winter, J.J. | | 登録日 | 2016-01-06 | | 公開日 | 2016-07-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | Mth1 Substrate Recognition--an Example of Specific Promiscuity.
Plos One, 11, 2016
|
|
8IQH
 
 | |
1RW2
 
 | | Three-dimensional structure of Ku80 CTD | | 分子名称: | ATP-dependent DNA helicase II, 80 kDa subunit | | 著者 | Zhang, Z, Hu, W, Cano, L, Lee, T.D, Chen, D.J, Chen, Y. | | 登録日 | 2003-12-15 | | 公開日 | 2003-12-30 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions.
STRUCTURE, 12, 2004
|
|
1TXI
 
 | | Crystal structure of the vdr ligand binding domain complexed to TX522 | | 分子名称: | (1R,3R)-5-((Z)-2-((1R,7AS)-HEXAHYDRO-1-((S)-6-HYDROXY-6-METHYLHEPT-4-YN-2-YL)-7A-METHYL-1H-INDEN-4(7AH)-YLIDENE)ETHYLIDENE)CYCLOHEXANE-1,3-DIOL, Vitamin D receptor | | 著者 | Moras, D, Rochel, N. | | 登録日 | 2004-07-05 | | 公開日 | 2005-05-10 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Superagonistic Action of 14-epi-Analogs of 1,25-Dihydroxyvitamin D Explained by Vitamin D Receptor-Coactivator Interaction
Mol.Pharmacol., 67, 2005
|
|
6JJ0
 
 | |
8A0A
 
 | |
6VI1
 
 | | Structure of Fab4 bound to P22 TerL(1-33) | | 分子名称: | Synthetic Fab4 heavy chain, Synthetic Fab4 light chain, Terminase, ... | | 著者 | Cingolani, G, Lokareddy, R, Ko, Y. | | 登録日 | 2020-01-11 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Recognition of an alpha-helical hairpin in P22 large terminase by a synthetic antibody fragment.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
4K4Y
 
 | | Coxsackievirus B3 polymerase elongation complex (r2+1_form) | | 分子名称: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, ACETATE ION, DNA/RNA (5'-R(*UP*GP*UP*UP*CP*GP*AP*CP*GP*AP*GP*AP*GP*AP*(DOC))-3'), ... | | 著者 | Gong, P, Peersen, O.B. | | 登録日 | 2013-04-12 | | 公開日 | 2013-05-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.72 Å) | | 主引用文献 | Structures of coxsackievirus, rhinovirus, and poliovirus polymerase elongation complexes solved by engineering RNA mediated crystal contacts.
Plos One, 8, 2013
|
|
6GTO
 
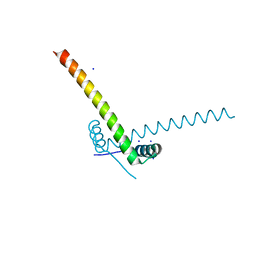 | | Structure of the AtaR antitoxin | | 分子名称: | DUF1778 domain-containing protein, SODIUM ION | | 著者 | Garcia-Pino, A, Jurenas, D. | | 登録日 | 2018-06-18 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.97 Å) | | 主引用文献 | Mechanism of regulation and neutralization of the AtaR-AtaT toxin-antitoxin system.
Nat. Chem. Biol., 15, 2019
|
|
1UHM
 
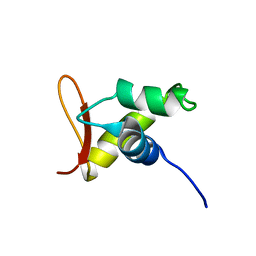 | | Solution structure of the globular domain of linker histone homolog Hho1p from S. cerevisiae | | 分子名称: | Histone H1 | | 著者 | Ono, K, Kusano, O, Shimotakahara, S, Shimizu, M, Yamazaki, T, Shindo, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2003-07-05 | | 公開日 | 2003-12-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The linker histone homolog Hho1p from Saccharomyces cerevisiae represents a winged helix-turn-helix fold as determined by NMR spectroscopy.
Nucleic Acids Res., 31, 2003
|
|
4HYY
 
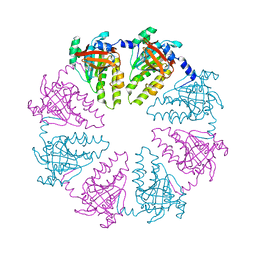 | |
6KI6
 
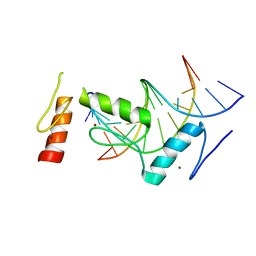 | | Crystal structure of BCL11A in complex with gamma-globin -115 HPFH region | | 分子名称: | B-cell lymphoma/leukemia 11A, DNA (5'-D(*AP*TP*AP*TP*TP*GP*GP*TP*CP*AP*AP*GP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*TP*GP*AP*CP*CP*AP*AP*TP*A)-3'), ... | | 著者 | Li, F.D, Yang, Y, Shi, Y.Y. | | 登録日 | 2019-07-17 | | 公開日 | 2019-09-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural insights into the recognition of gamma-globin gene promoter by BCL11A.
Cell Res., 29, 2019
|
|
1A63
 
 | | THE NMR STRUCTURE OF THE RNA BINDING DOMAIN OF E.COLI RHO FACTOR SUGGESTS POSSIBLE RNA-PROTEIN INTERACTIONS, 10 STRUCTURES | | 分子名称: | RHO | | 著者 | Briercheck, D.M, Wood, T.C, Allison, T.J, Richardson, J.P, Rule, G.S. | | 登録日 | 1998-03-05 | | 公開日 | 1998-05-27 | | 最終更新日 | 2024-04-10 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The NMR structure of the RNA binding domain of E. coli rho factor suggests possible RNA-protein interactions.
Nat.Struct.Biol., 5, 1998
|
|
1UVJ
 
 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 with 7nt RNA | | 分子名称: | 5'-R(*UP*UP*CP*CP)-3', MANGANESE (II) ION, RNA-directed RNA polymerase | | 著者 | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | 登録日 | 2004-01-21 | | 公開日 | 2004-02-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
4E1N
 
 | |
3K18
 
 | |
