7ONB
 
 | |
7XYG
 
 | |
3GSZ
 
 | |
2E1W
 
 | |
8HXX
 
 | |
7XYF
 
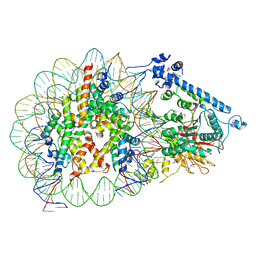 | |
4UTP
 
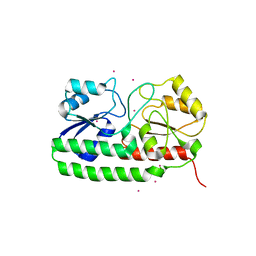 | | Crystal structure of pneumococcal surface antigen PsaA in the Cd- bound, closed state | | 分子名称: | CADMIUM ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | 著者 | Luo, Z, Counago, R.M, Maher, M, Kobe, B. | | 登録日 | 2014-07-22 | | 公開日 | 2014-08-13 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae.
Nat Commun, 6, 2015
|
|
4UTO
 
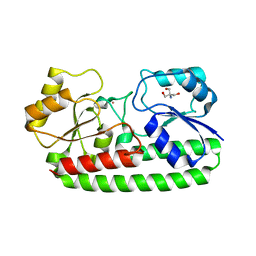 | | Crystal structure of pneumococcal surface antigen PsaA D280N in the Cd-bound, open state | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, MANGANESE ABC TRANSPORTER SUBSTRATE-BINDING LIPOPROTEIN | | 著者 | Luo, Z, Counago, R.M, Maher, M, Kobe, B. | | 登録日 | 2014-07-22 | | 公開日 | 2015-03-11 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Dysregulation of transition metal ion homeostasis is the molecular basis for cadmium toxicity in Streptococcus pneumoniae.
Nat Commun, 6, 2015
|
|
7YRD
 
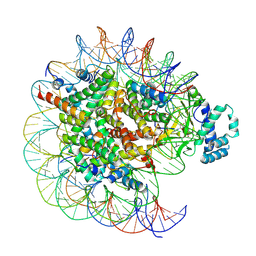 | | histone methyltransferase | | 分子名称: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | 著者 | Li, H, Wang, W.Y. | | 登録日 | 2022-08-09 | | 公開日 | 2023-08-16 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
1HNI
 
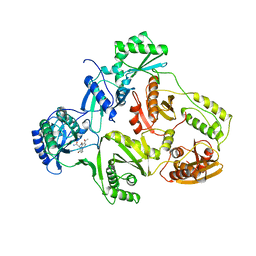 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN A COMPLEX WITH THE NONNUCLEOSIDE INHIBITOR ALPHA-APA R 95845 AT 2.8 ANGSTROMS RESOLUTION | | 分子名称: | (2-ACETYL-5-METHYLANILINO)(2,6-DIBROMOPHENYL)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | 著者 | Ding, J, Das, K, Arnold, E. | | 登録日 | 1995-02-28 | | 公開日 | 1995-06-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of HIV-1 reverse transcriptase in a complex with the non-nucleoside inhibitor alpha-APA R 95845 at 2.8 A resolution.
Structure, 3, 1995
|
|
2GUT
 
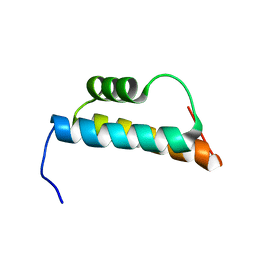 | | Solution structure of the trans-activation domain of the human co-activator ARC105 | | 分子名称: | ARC/MEDIATOR, Positive cofactor 2 glutamine/Q-rich-associated protein | | 著者 | Vought, B.W, Jim Sun, Z.-Y, Hyberts, S.G, Wagner, G, Naar, A.M. | | 登録日 | 2006-05-01 | | 公開日 | 2006-08-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | An ARC/Mediator subunit required for SREBP control of cholesterol and lipid homeostasis.
Nature, 442, 2006
|
|
1KV7
 
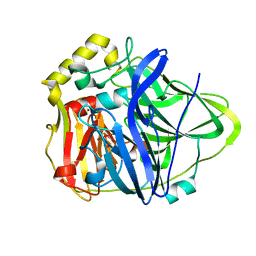 | | Crystal Structure of CueO, a multi-copper oxidase from E. coli involved in copper homeostasis | | 分子名称: | COPPER (II) ION, CU-O-CU LINKAGE, PROBABLE BLUE-COPPER PROTEIN YACK | | 著者 | Roberts, S.A, Weichsel, A, Grass, G, Thakali, K, Hazzard, J.T, Tollin, G, Rensing, C, Montfort, W.R. | | 登録日 | 2002-01-25 | | 公開日 | 2002-02-06 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
6TJ6
 
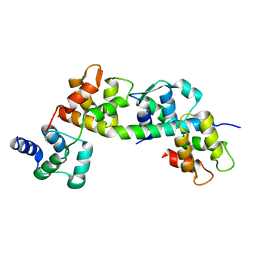 | |
6TJ5
 
 | |
6TJ4
 
 | |
1QXL
 
 | | Crystal structure of Adenosine deaminase complexed with FR235380 | | 分子名称: | 1-((1R)-1-(HYDROXYMETHYL)-3-{6-[(5-PHENYLPENTANOYL)AMINO]-1H-INDOL-1-YL}PROPYL)-1H-IMIDAZOLE-4-CARBOXAMIDE, Adenosine deaminase, ZINC ION | | 著者 | Kinoshita, T. | | 登録日 | 2003-09-08 | | 公開日 | 2004-09-21 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structure-based design, synthesis, and structure-activity relationship studies of novel non-nucleoside adenosine deaminase inhibitors
J.Med.Chem., 47, 2004
|
|
6LTH
 
 | | Structure of human BAF Base module | | 分子名称: | AT-rich interactive domain-containing protein 1A, SWI/SNF complex subunit SMARCC2, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1, ... | | 著者 | He, S, Wu, Z, Tian, Y, Yu, Z, Yu, J, Wang, X, Li, J, Liu, B, Xu, Y. | | 登録日 | 2020-01-22 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of nucleosome-bound human BAF complex.
Science, 367, 2020
|
|
7YRG
 
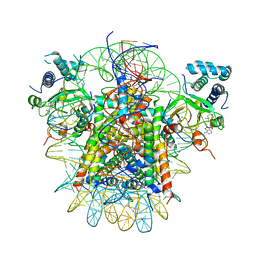 | | histone methyltransferase | | 分子名称: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | 著者 | Li, H, Wang, W.Y. | | 登録日 | 2022-08-09 | | 公開日 | 2023-12-13 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
1A4M
 
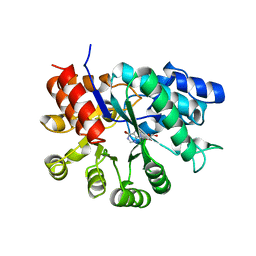 | | ADA STRUCTURE COMPLEXED WITH PURINE RIBOSIDE AT PH 7.0 | | 分子名称: | 6-HYDROXY-1,6-DIHYDRO PURINE NUCLEOSIDE, ADENOSINE DEAMINASE, ZINC ION | | 著者 | Wang, Z, Quiocho, F.A. | | 登録日 | 1998-01-31 | | 公開日 | 1998-10-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Complexes of adenosine deaminase with two potent inhibitors: X-ray structures in four independent molecules at pH of maximum activity.
Biochemistry, 37, 1998
|
|
6WWD
 
 | |
4J06
 
 | |
4J02
 
 | | Crystal structure of hcv ns5b polymerase in complex with [(1R)-5,8-DICHLORO-1-PROPYL-1,3,4,9-TETRAHYDROPYRANO[3,4-B]INDOL-1-YL]ACETIC ACID | | 分子名称: | Genome polyprotein, SODIUM ION, SULFATE ION, ... | | 著者 | Coulombe, R. | | 登録日 | 2013-01-30 | | 公開日 | 2013-04-17 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery of a novel series of non-nucleoside thumb pocket 2 HCV NS5B polymerase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4IZ0
 
 | | Crystal structure of HCV NS5B polymerase in complex with 2,4,5-trichloro-N-(5-methyl-1,2-oxazol-3-yl)benzenesulfonamide | | 分子名称: | 2,4,5-trichloro-N-(5-methyl-1,2-oxazol-3-yl)benzenesulfonamide, RNA-directed RNA polymerase | | 著者 | Coulombe, R. | | 登録日 | 2013-01-29 | | 公開日 | 2013-04-17 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.22 Å) | | 主引用文献 | Discovery of a novel series of non-nucleoside thumb pocket 2 HCV NS5B polymerase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4J04
 
 | |
4J08
 
 | |
