3GPW
 
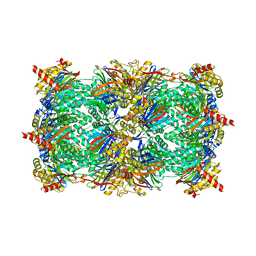 | | Crystal structure of the yeast 20S proteasome in complex with Salinosporamide derivatives: irreversible inhibitor ligand | | 分子名称: | (3AR,6R,6AS)-6-((S)-((S)-CYCLOHEX-2-ENYL)(HYDROXY)METHYL)-6A-METHYL-4-OXO-HEXAHYDRO-2H-FURO[3,2-C]PYRROLE-6-CARBALDEHYDE, Proteasome component C1, Proteasome component C11, ... | | 著者 | Groll, M, Macherla, V.R, Manam, R.R, Arthur, K.A.M, Potts, C.B. | | 登録日 | 2009-03-23 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Snapshots of the fluorosalinosporamide/20S complex offer mechanistic insights for fine tuning proteasome inhibition
J.Med.Chem., 52, 2009
|
|
1Q6Z
 
 | |
3GXM
 
 | |
3H44
 
 | |
3GPT
 
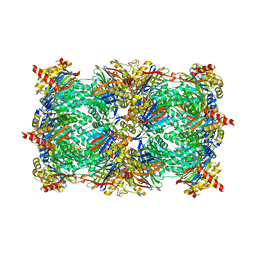 | | Crystal structure of the yeast 20S proteasome in complex with Salinosporamide derivatives: slow substrate ligand | | 分子名称: | (2R,3S,4R)-2-[(S)-(1S)-cyclohex-2-en-1-yl(hydroxy)methyl]-4-(2-fluoroethyl)-3-hydroxy-3-methyl-5-oxopyrrolidine-2-carbaldehyde, Proteasome component C1, Proteasome component C11, ... | | 著者 | Groll, M, Macherla, V.R, Manam, R.R, Arthur, K.A.M, Potts, C.B. | | 登録日 | 2009-03-23 | | 公開日 | 2009-09-15 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Snapshots of the fluorosalinosporamide/20S complex offer mechanistic insights for fine tuning proteasome inhibition
J.Med.Chem., 52, 2009
|
|
3H6H
 
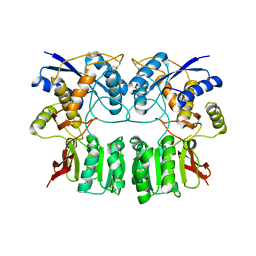 | |
1QYE
 
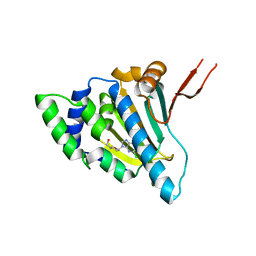 | | Crystal Structure of the N-domain of the ER Hsp90 chaperone GRP94 in complex with 2-chlorodideoxyadenosine | | 分子名称: | 1-METHOXY-2-(2-METHOXYETHOXY)ETHANE, 2-CHLORODIDEOXYADENOSINE, Endoplasmin | | 著者 | Soldano, K.L, Jivan, A, Nicchitta, C.V, Gewirth, D.T. | | 登録日 | 2003-09-10 | | 公開日 | 2003-11-11 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structure of the N-terminal Domain of GRP94. Basis for Ligand Specificity and Regulation
J.Biol.Chem., 278, 2003
|
|
1QZ1
 
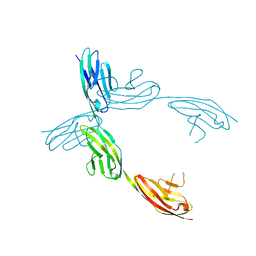 | | Crystal Structure of the Ig 1-2-3 fragment of NCAM | | 分子名称: | Neural cell adhesion molecule 1, 140 kDa isoform | | 著者 | Soroka, V, Kolkova, K, Kastrup, J.S, Diederichs, K, Breed, J, Kiselyov, V.V, Poulsen, F.M, Larsen, I.K, Welte, W, Berezin, V, Bock, E, Kasper, C. | | 登録日 | 2003-09-15 | | 公開日 | 2003-11-04 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structure and interactions of NCAM Ig1-2-3 suggest a novel zipper mechanism for homophilic adhesion
Structure, 11, 2003
|
|
3HC6
 
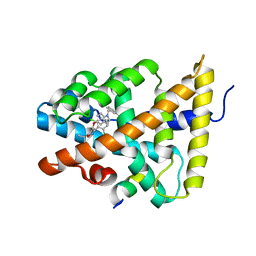 | | FXR with SRC1 and GSK088 | | 分子名称: | 3-[(5-{[3-(2,6-dichlorophenyl)-5-(1-methylethyl)isoxazol-4-yl]methoxy}-1H-indol-1-yl)methyl]benzoic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | 著者 | Williams, S.P, Madauss, K.P. | | 登録日 | 2009-05-05 | | 公開日 | 2009-07-21 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | FXR agonist activity of conformationally constrained analogs of GW 4064.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1R0A
 
 | | Crystal structure of HIV-1 reverse transcriptase covalently tethered to DNA template-primer solved to 2.8 angstroms | | 分子名称: | 5'-D(*A*TP*GP*CP*AP*TP*CP*GP*GP*CP*GP*CP*TP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*GP*GP*T)-3', 5'-D(*C*CP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*AP*GP*CP*GP*CP*CP*GP*(2DA))-3', GLYCEROL, ... | | 著者 | Tuske, S, Ding, J, Arnold, E. | | 登録日 | 2003-09-19 | | 公開日 | 2004-08-03 | | 最終更新日 | 2021-10-27 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Nonnucleoside inhibitor binding affects the interactions of the fingers subdomain of human immunodeficiency virus type 1 reverse transcriptase with DNA.
J.Virol., 78, 2004
|
|
1QYF
 
 | | Crystal structure of matured green fluorescent protein R96A variant | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, green-fluorescent protein | | 著者 | Barondeau, D.P, Putnam, C.D, Kassmann, C.J, Tainer, J.A, Getzoff, E.D. | | 登録日 | 2003-09-10 | | 公開日 | 2003-09-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Mechanism and energetics of green fluorescent protein chromophore synthesis revealed by trapped intermediate structures.
Proc.Natl.Acad.Sci.Usa, 100, 2003
|
|
3HE6
 
 | |
3HF2
 
 | | Crystal structure of the I401P mutant of cytochrome P450 BM3 | | 分子名称: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Yang, W, Whitehouse, C.J.C, Bell, S.G, Bartlam, M, Wong, L.L, Rao, Z. | | 登録日 | 2009-05-10 | | 公開日 | 2009-06-30 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | A Highly Active Single-Mutation Variant of P450(BM3) (CYP102A1)
Chembiochem, 10, 2009
|
|
3HF9
 
 | |
1R05
 
 | | Solution Structure of Max B-HLH-LZ | | 分子名称: | Max protein | | 著者 | Sauv, S, Tremblay, L, Lavigne, P. | | 登録日 | 2003-09-19 | | 公開日 | 2003-10-21 | | 最終更新日 | 2021-10-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The NMR solution structure of a mutant of the Max b/HLH/LZ free of DNA: insights into the specific and reversible DNA binding mechanism of dimeric transcription factors
J.Mol.Biol., 342, 2004
|
|
3LFI
 
 | |
4BJ1
 
 | | Crystal structure of Saccharomyces cerevisiae RIF2 | | 分子名称: | CHLORIDE ION, PROTEIN RIF2 | | 著者 | Shi, T, Bunker, R.D, Gut, H, Scrima, A, Thoma, N.H. | | 登録日 | 2013-04-15 | | 公開日 | 2013-06-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Rif1 and Rif2 Shape Telomere Funcation and Architecture Through Multivalent RAP1 Interactions
Cell(Cambridge,Mass.), 153, 2013
|
|
6ZC7
 
 | |
4BLD
 
 | | Crystal structure of a human Suppressor of fused (SUFU)-GLI3p complex | | 分子名称: | MALTOSE-BINDING PERIPLASMIC PROTEIN, SUPPRESSOR OF FUSED HOMOLOG, TRANSCRIPTIONAL ACTIVATOR GLI3, ... | | 著者 | Cherry, A.L, Finta, C, Karlstrom, M, De Sanctis, D, Toftgard, R, Jovine, L. | | 登録日 | 2013-05-02 | | 公開日 | 2013-11-27 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.802 Å) | | 主引用文献 | Structural Basis of Sufu-GLI Interaction in Hedgehog Signalling Regulation
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3LTE
 
 | | CRYSTAL STRUCTURE OF RESPONSE REGULATOR (SIGNAL RECEIVER DOMAIN) FROM Bermanella marisrubri | | 分子名称: | GLYCEROL, PHOSPHATE ION, Response regulator | | 著者 | Patskovsky, Y, Toro, R, Gilmore, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2010-02-15 | | 公開日 | 2010-03-31 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | CRYSTAL STRUCTURE OF RESPONSE REGULATOR SIGNAL RECEIVER DOMAIN FROM Bermanella marisrubri RED65
To be Published
|
|
3IG9
 
 | | Small outer capsid protein (SOC) of bacteriophage RB69 | | 分子名称: | IMIDAZOLE, Soc small outer capsid protein | | 著者 | Li, Q, Fokine, A, O'Donnell, E, Rao, V.B, Rossmann, M.G. | | 登録日 | 2009-07-27 | | 公開日 | 2009-12-08 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of the Small Outer Capsid Protein, Soc: A Clamp for Stabilizing Capsids of T4-like Phages
J.Mol.Biol., 395, 2010
|
|
5LF0
 
 | | Human 20S proteasome complex with Epoxomicin at 2.4 Angstrom | | 分子名称: | CHLORIDE ION, EPOXOMICIN (peptide inhibitor), MAGNESIUM ION, ... | | 著者 | Schrader, J, Henneberg, F, Mata, R, Tittmann, K, Schneider, T.R, Stark, H, Bourenkov, G, Chari, A. | | 登録日 | 2016-06-30 | | 公開日 | 2016-08-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | The inhibition mechanism of human 20S proteasomes enables next-generation inhibitor design.
Science, 353, 2016
|
|
3M32
 
 | | Structural Insight into Methyl-Coenzyme M Reductase Chemistry using Coenzyme B Analogues | | 分子名称: | 1,2-ETHANEDIOL, 1-THIOETHANESULFONIC ACID, ACETATE ION, ... | | 著者 | Cedervall, P.E, Dey, M, Ragsdale, S.W, Wilmot, C.M. | | 登録日 | 2010-03-08 | | 公開日 | 2010-09-15 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural insight into methyl-coenzyme M reductase chemistry using coenzyme B analogues.
Biochemistry, 49, 2010
|
|
4BEW
 
 | | SERCA bound to phosphate analogue | | 分子名称: | (6AR,11AS,11BR)-10-ACETYL-9-HYDROXY-7,7-DIMETHYL-2,6,6A,7,11A,11B-HEXAHYDRO-11H-PYRROLO[1',2':2,3]ISOINDOLO[4,5,6-CD]INDOL-11-ONE, ACETATE ION, MAGNESIUM ION, ... | | 著者 | Drachmann, N.D, Mattle, D, Laursen, M, Bublitz, M, Olesen, C, Moeller, J.V, Nissen, P, Morth, J.P. | | 登録日 | 2013-03-12 | | 公開日 | 2014-02-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Serca Bound to Phosphate Analogue
To be Published
|
|
3LK0
 
 | | X-ray structure of bovine SC0067,Ca(2+)-S100B | | 分子名称: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, CALCIUM ION, Protein S100-B | | 著者 | Charpentier, T.H, Weber, D.J, Wilder, P.W. | | 登録日 | 2010-01-26 | | 公開日 | 2010-12-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | In vitro screening and structural characterization of inhibitors of the S100B-p53 interaction.
Int J High Throughput Screen, 2010, 2010
|
|
