4EPQ
 
 | |
4JWR
 
 | |
3O80
 
 | | Crystal structure of monomeric KlHxk1 in crystal form IX (open state) | | 分子名称: | Hexokinase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Kuettner, E.B, Kettner, K, Keim, A, Kriegel, T.M, Strater, N. | | 登録日 | 2010-08-02 | | 公開日 | 2010-10-13 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Crystal Structure of Hexokinase KlHxk1 of Kluyveromyces lactis: A MOLECULAR BASIS FOR UNDERSTANDING THE CONTROL OF YEAST HEXOKINASE FUNCTIONS VIA COVALENT MODIFICATION AND OLIGOMERIZATION.
J.Biol.Chem., 285, 2010
|
|
3O72
 
 | | Crystal structure of EfeB in complex with heme | | 分子名称: | OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, Redox component of a tripartite ferrous iron transporter | | 著者 | Liu, X, Du, Q, Wang, Z, Zhu, D, Huang, Y, Li, N, Xu, S, Gu, L. | | 登録日 | 2010-07-30 | | 公開日 | 2011-03-16 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure and biochemical features of EfeB/YcdB from Escherichia coli O157: ASP235 plays divergent roles in different enzyme-catalyzed processes
J.Biol.Chem., 286, 2011
|
|
3QQ7
 
 | |
4FDD
 
 | | Crystal structure of KAP beta2-PY-NLS | | 分子名称: | RNA-binding protein FUS, Transportin-1 | | 著者 | Zhang, Z.C, Chook, Y.M. | | 登録日 | 2012-05-28 | | 公開日 | 2012-07-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and energetic basis of ALS-causing mutations in the atypical proline-tyrosine nuclear localization signal of the Fused in Sarcoma protein (FUS).
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FMJ
 
 | | Merkel cell polyomavirus VP1 in complex with GD1a oligosaccharide | | 分子名称: | CHLORIDE ION, GLYCEROL, N-acetyl-alpha-neuraminic acid, ... | | 著者 | Neu, U, Hengel, H, Stehle, T. | | 登録日 | 2012-06-17 | | 公開日 | 2012-09-05 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structures of Merkel Cell Polyomavirus VP1 Complexes Define a Sialic Acid Binding Site Required for Infection.
Plos Pathog., 8, 2012
|
|
3U6S
 
 | | MutM set 1 TpG | | 分子名称: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*TP*CP*CP*AP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*TP*GP*GP*AP*(08Q)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | 著者 | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | 登録日 | 2011-10-12 | | 公開日 | 2012-04-25 | | 最終更新日 | 2013-09-25 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
3U80
 
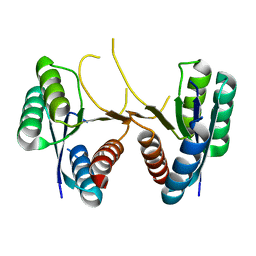 | | 1.60 Angstrom Resolution Crystal Structure of a 3-Dehydroquinate Dehydratase-like Protein from Bifidobacterium longum | | 分子名称: | 3-dehydroquinate dehydratase, type II | | 著者 | Light, S.H, Minasov, G, Shuvalova, L, Papazisi, L, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2011-10-14 | | 公開日 | 2011-10-26 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal structure of a type II dehydroquinate dehydratase-like protein from Bifidobacterium longum.
J.Struct.Funct.Genom., 14, 2013
|
|
4LFT
 
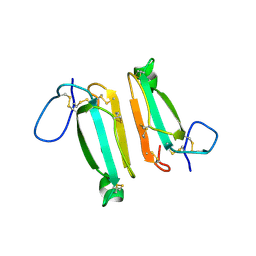 | | Structure of alpha-elapitoxin-Dpp2d isolated from Black Mamba (Dendroaspis polylepis) venom | | 分子名称: | Alpha-elapitoxin-Dpp2a | | 著者 | Wang, C.I.A, Reeks, T, Lewis, R.J, Alewood, P.F, Durek, T. | | 登録日 | 2013-06-27 | | 公開日 | 2014-06-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Isolation and Structural and Pharmacological Characterization of alpha-Elapitoxin-Dpp2d, an Amidated Three Finger Toxin from Black Mamba Venom.
Biochemistry, 53, 2014
|
|
3QUX
 
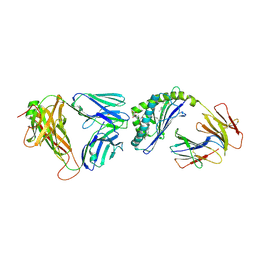 | | Structure of the mouse CD1d-alpha-C-GalCer-iNKT TCR complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | 著者 | Li, Y, Girardi, E, Yu, E.D, Zajonc, D.M. | | 登録日 | 2011-02-24 | | 公開日 | 2011-06-29 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Galactose-modified iNKT cell agonists stabilized by an induced fit of CD1d prevent tumour metastasis.
Embo J., 30, 2011
|
|
4L6A
 
 | | Structure of human mitochondrial 5'(3')-deoxyribonucleotidase | | 分子名称: | 5'(3')-deoxyribonucleotidase, mitochondrial, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Pachl, P, Brynda, J, Rezacova, P. | | 登録日 | 2013-06-12 | | 公開日 | 2014-02-12 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structures of human cytosolic and mitochondrial nucleotidases: implications for structure-based design of selective inhibitors.
Acta Crystallogr. D Biol. Crystallogr., 70, 2014
|
|
3U6D
 
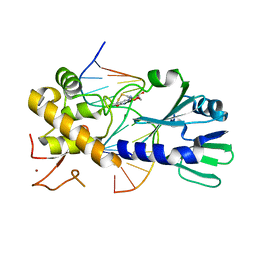 | | MutM set 1 GpGo | | 分子名称: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*TP*CP*CP*CP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*GP*(8OG)P*GP*AP*(TX)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | 著者 | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | 登録日 | 2011-10-12 | | 公開日 | 2012-04-25 | | 最終更新日 | 2013-09-25 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
4HBV
 
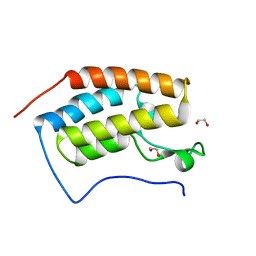 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolin ligand | | 分子名称: | 1,2-ETHANEDIOL, 6-bromo-3-methyl-3,4-dihydroquinazolin-2(1H)-one, Bromodomain-containing protein 4 | | 著者 | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2012-09-28 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
4HBY
 
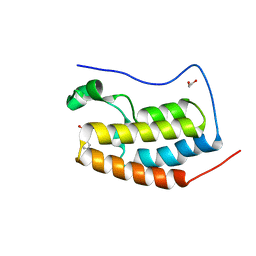 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a quinazolin ligand | | 分子名称: | 1,2-ETHANEDIOL, 3-methyl-2-oxo-N-phenyl-1,2,3,4-tetrahydroquinazoline-6-sulfonamide, Bromodomain-containing protein 4 | | 著者 | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Fish, P.V, Bunnage, M.E, Cook, A.S, Owen, D.R, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2012-09-28 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Identification of a Chemical Probe for Bromo and Extra C-Terminal Bromodomain Inhibition through Optimization of a Fragment-Derived Hit.
J.Med.Chem., 55, 2012
|
|
3OWN
 
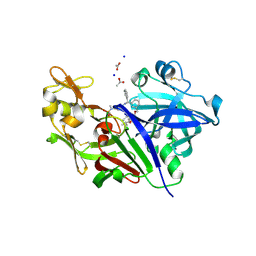 | | Potent macrocyclic renin inhibitors | | 分子名称: | (2S,4S)-4-hydroxy-2-(1-methylethyl)-4-[(4R,13S)-18-[methyl(methylsulfonyl)amino]-2,15-dioxo-4-phenyl-11-oxa-3,14-diazatricyclo[14.3.1.1~5,9~]henicosa-1(20),5(21),6,8,16,18-hexaen-13-yl]-N-(2-methylpropyl)butanamide, (2S,4S)-4-hydroxy-2-(1-methylethyl)-4-[(4S,13S)-18-[methyl(methylsulfonyl)amino]-2,15-dioxo-4-phenyl-11-oxa-3,14-diazatricyclo[14.3.1.1~5,9~]henicosa-1(20),5(21),6,8,16,18-hexaen-13-yl]-N-(2-methylpropyl)butanamide, ACETATE ION, ... | | 著者 | Borkakoti, N, Derbyshire, D. | | 登録日 | 2010-09-20 | | 公開日 | 2010-12-15 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Design and synthesis of potent macrocyclic renin inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4GUS
 
 | | Crystal structure of LSD2-NPAC with H3 in space group P3221 | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Histone H3.3, ... | | 著者 | Chen, F, Dong, Z, Fang, J, Yang, Y, Li, Z, Xu, Y, Yang, H, Wang, P, Fang, R, Shi, Y, Xu, Y. | | 登録日 | 2012-08-29 | | 公開日 | 2013-01-16 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Mol.Cell, 49, 2013
|
|
3P9K
 
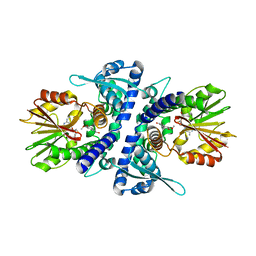 | |
4JPE
 
 | | Spirocyclic Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors | | 分子名称: | (4R)-2-amino-1,3',3'-trimethyl-7'-(pyrimidin-5-yl)-3',4'-dihydro-2'H-spiro[imidazole-4,1'-naphthalen]-5(1H)-one, Beta-secretase 1, NICKEL (II) ION | | 著者 | Vigers, G.P.A, Smith, D. | | 登録日 | 2013-03-19 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Spirocyclic beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors: from hit to lowering of cerebrospinal fluid (CSF) amyloid beta in a higher species.
J.Med.Chem., 56, 2013
|
|
4KVX
 
 | |
4FK6
 
 | | JAK1 kinase (JH1 domain) in complex with compound 72 | | 分子名称: | N-({1-[(1R,2R,4S)-bicyclo[2.2.1]hept-2-yl]-1,6-dihydroimidazo[4,5-d]pyrrolo[2,3-b]pyridin-2-yl}methyl)methanesulfonamide, Tyrosine-protein kinase JAK1 | | 著者 | Eigenbrot, C, Steffek, M. | | 登録日 | 2012-06-12 | | 公開日 | 2012-11-07 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure-based discovery of C-2 substituted imidazo-pyrrolopyridine JAK1 inhibitors with improved selectivity over JAK2.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4KXT
 
 | |
3PJI
 
 | |
4L0L
 
 | | Crystal structure of P.aeruginosa PBP3 in complex with compound 4 | | 分子名称: | (6R,7S,10Z)-10-(2-amino-1,3-thiazol-4-yl)-1-(1,5-dihydroxy-4-oxo-1,4-dihydropyridin-2-yl)-7-formyl-13,13-dimethyl-3,9-dioxo-6-(sulfoamino)-12-oxa-2,4,8,11-tetraazatetradec-10-en-14-oic acid, Penicillin-binding protein 3 | | 著者 | Han, S, Marr, E.S. | | 登録日 | 2013-05-31 | | 公開日 | 2013-08-21 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Pyridone-conjugated monobactam antibiotics with gram-negative activity.
J.Med.Chem., 56, 2013
|
|
4L1G
 
 | |
