4BS7
 
 | | Hen egg-white lysozyme structure determined at room temperature by in- situ diffraction and SAD phasing in ChipX | | 分子名称: | LYSOZYME C, YTTERBIUM (III) ION | | 著者 | Pinker, F, Lorber, B, Olieric, V, Sauter, C. | | 登録日 | 2013-06-07 | | 公開日 | 2013-10-30 | | 最終更新日 | 2019-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.701 Å) | | 主引用文献 | Chipx: A Novel Microfluidic Chip for Counter- Diffusion Crystallization of Biomolecules and in Situ Crystal Analysis at Room Temperature
Cryst.Growth Des., 13, 2013
|
|
3MDM
 
 | | Thioperamide complex of Cytochrome P450 46A1 | | 分子名称: | Cholesterol 24-hydroxylase, N-cyclohexyl-4-(1H-imidazol-5-yl)piperidine-1-carbothioamide, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Mast, N, Charvet, C, Pikuleva, I, Stout, C.D. | | 登録日 | 2010-03-30 | | 公開日 | 2010-07-28 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain.
J.Biol.Chem., 285, 2010
|
|
6EOF
 
 | | Crystal structure of AMPylated GRP78 in ADP state | | 分子名称: | 78 kDa glucose-regulated protein, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | 著者 | Yan, Y, Preissler, S, Read, R.J, Ron, D. | | 登録日 | 2017-10-09 | | 公開日 | 2017-11-01 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
4BUO
 
 | | High Resolution Structure of Thermostable Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | 分子名称: | GLYCINE, NEUROTENSIN RECEPTOR TYPE 1, NEUROTENSIN/NEUROMEDIN N | | 著者 | Egloff, P, Hillenbrand, M, Schlinkmann, K.M, Batyuk, A, Mittl, P, Plueckthun, A. | | 登録日 | 2013-06-21 | | 公開日 | 2014-01-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6ES2
 
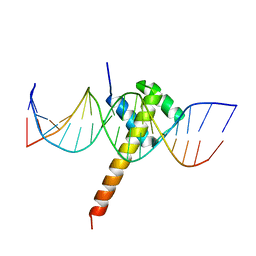 | | Structure of CDX2-DNA(CAA) | | 分子名称: | DNA (5'-D(P*GP*GP*AP*GP*GP*CP*AP*AP*TP*AP*AP*AP*AP*CP*AP*CP*AP*A)-3'), DNA (5'-D(P*TP*TP*GP*TP*GP*TP*TP*TP*TP*AP*TP*TP*GP*CP*CP*TP*CP*C)-3'), Homeobox protein CDX-2 | | 著者 | Morgunova, E, Yin, Y, Jolma, A, Popov, A, Taipale, J. | | 登録日 | 2017-10-19 | | 公開日 | 2018-03-21 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Two distinct DNA sequences recognized by transcription factors represent enthalpy and entropy optima.
Elife, 7, 2018
|
|
6E6C
 
 | | HRAS G13D bound to GppNHp (H13GNP) | | 分子名称: | CALCIUM ION, GTPase HRas, MAGNESIUM ION, ... | | 著者 | Johnson, C.W, Mattos, C. | | 登録日 | 2018-07-24 | | 公開日 | 2019-07-31 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D.
Cell Rep, 28, 2019
|
|
6E6H
 
 | | NRAS G13D bound to GppNHp (N13GNP) | | 分子名称: | GTPase NRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Johnson, C.W, Mattos, C. | | 登録日 | 2018-07-24 | | 公開日 | 2019-07-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Isoform-Specific Destabilization of the Active Site Reveals a Molecular Mechanism of Intrinsic Activation of KRas G13D.
Cell Rep, 28, 2019
|
|
6E6L
 
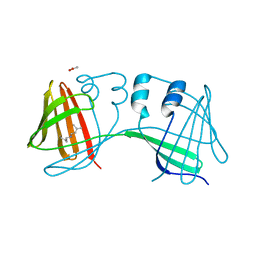 | |
3MHW
 
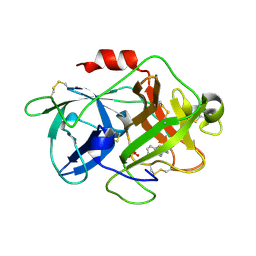 | | The complex crystal Structure of Urokianse and 2-Aminobenzothiazole | | 分子名称: | 1,3-benzothiazol-2-amine, SULFATE ION, Urokinase-type plasminogen activator | | 著者 | Jiang, L.-G, Yuan, C, Chen, L.-Q, Huang, M.-D. | | 登録日 | 2010-04-09 | | 公開日 | 2010-04-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Crystal Structures of 2-Aminobenzothiazole-based Inhibitors in Complexes with Urokinase-type Plasminogen Activator
CHIN.J.STRUCT.CHEM., 28, 2009
|
|
4CAJ
 
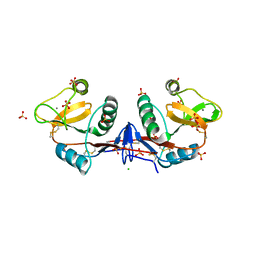 | | Crystallographic structure of the mouse SIGN-R1 CRD domain in complex with sialic acid | | 分子名称: | CALCIUM ION, CD209 ANTIGEN-LIKE PROTEIN B, CHLORIDE ION, ... | | 著者 | Silva-Martin, N, Bartual, S.G, Hermoso, J.A. | | 登録日 | 2013-10-08 | | 公開日 | 2014-10-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.191 Å) | | 主引用文献 | Structural Basis for Selective Recognition of Endogenous and Microbial Polysaccharides by Macrophage Receptor Sign-R1
Structure, 22, 2014
|
|
6EAU
 
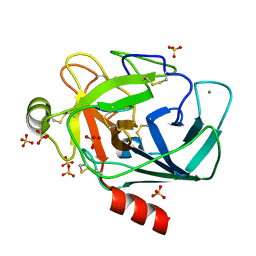 | | Crystallographic structure of the octapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 21 21 21. | | 分子名称: | CALCIUM ION, CYS-THR-LYS-SER-ILE-PRO-PRO-CYS, Cationic trypsin, ... | | 著者 | Fernandes, J.C, Valadares, N.F, Freitas, S.M, Barbosa, J.A.R.G. | | 登録日 | 2018-08-03 | | 公開日 | 2019-08-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.18 Å) | | 主引用文献 | Crystallographic structure of the octapeptide derived from the BTCI inhibitor bound to beta-trypsin in space group P 21 21 21.
To Be Published
|
|
3MN6
 
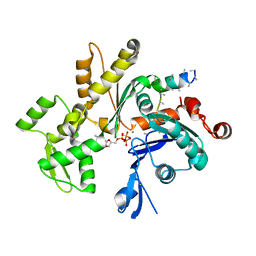 | | Structures of actin-bound WH2 domains of Spire and the implication for filament nucleation | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION, ... | | 著者 | Ducka, A.M, Sitar, T, Popowicz, G.M, Huber, R, Holak, T.A. | | 登録日 | 2010-04-21 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4BQC
 
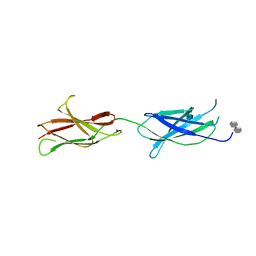 | | Crystal structure of the FN5 and FN6 domains of NEO1 bound to SOS | | 分子名称: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, NEOGENIN, ... | | 著者 | Bell, C.H, Healey, E, vanErp, S, Bishop, B, Tang, C, Gilbert, R.J.C, Aricescu, A.R, Pasterkamp, R.J, Siebold, C. | | 登録日 | 2013-05-30 | | 公開日 | 2013-06-12 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of the Repulsive Guidance Molecule (Rgm)-Neogenin Signaling Hub
Science, 341, 2013
|
|
3LZS
 
 | | Crystal Structure of HIV-1 CRF01_AE Protease in Complex with Darunavir | | 分子名称: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, ACETATE ION, HIV-1 protease | | 著者 | Schiffer, C.A, Bandaranayake, R.M. | | 登録日 | 2010-03-01 | | 公開日 | 2010-08-11 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The Effect of Clade-Specific Sequence Polymorphisms on HIV-1 Protease Activity and Inhibitor Resistance Pathways.
J.Virol., 84, 2010
|
|
3M35
 
 | |
3M3B
 
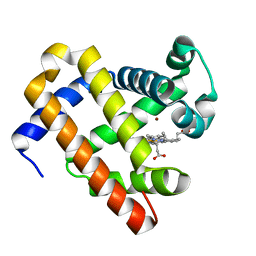 | | The roles of glutamates and metal ions in a rationally designed nitric oxide reductase based on myoglobin: Zn(II)-I107E FeBMb (Zn(II) binding to FeB site) | | 分子名称: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ZINC ION | | 著者 | Lin, Y.-W, Yeung, N, Gao, Y.-G, Miner, K.D, Tian, S, Robinson, H, Lu, Y. | | 登録日 | 2010-03-08 | | 公開日 | 2010-05-12 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Roles of glutamates and metal ions in a rationally designed nitric oxide reductase based on myoglobin.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4BVV
 
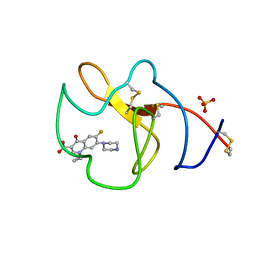 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | 分子名称: | 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, APOLIPOPROTEIN(A), SULFATE ION | | 著者 | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | 登録日 | 2013-06-28 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
3M4V
 
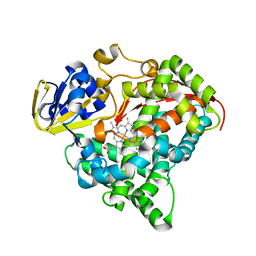 | | Crystal structure of the A330P mutant of cytochrome P450 BM3 | | 分子名称: | Bifunctional P-450/NADPH-P450 reductase, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Yang, W, Whitehouse, C.J.C, Bell, S.G, Bartlam, M, Wong, L.L, Rao, Z. | | 登録日 | 2010-03-12 | | 公開日 | 2011-03-23 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural basis for the properties of two single-site proline mutants of CYP102A1 (P450BM3)
Chembiochem, 11, 2010
|
|
6EQM
 
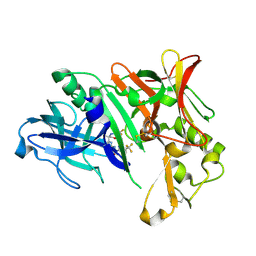 | | Crystal Structure of Human BACE-1 in Complex with CNP520 | | 分子名称: | Beta-secretase 1, ~{N}-[6-[(3~{R},6~{R})-5-azanyl-3,6-dimethyl-6-(trifluoromethyl)-2~{H}-1,4-oxazin-3-yl]-5-fluoranyl-pyridin-2-yl]-3-chloranyl-5-(trifluoromethyl)pyridine-2-carboxamide | | 著者 | Rondeau, J.-M, Wirth, E. | | 登録日 | 2017-10-13 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | The BACE-1 inhibitor CNP520 for prevention trials in Alzheimer's disease.
EMBO Mol Med, 10, 2018
|
|
3MN9
 
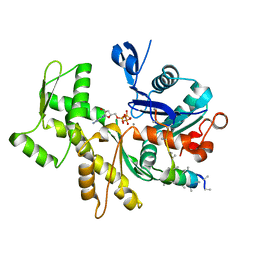 | | Structures of actin-bound WH2 domains of Spire and the implication for filament nucleation | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Actin-5C, CALCIUM ION, ... | | 著者 | Ducka, A.M, Sitar, T, Popowicz, G.M, Huber, R, Holak, T.A. | | 登録日 | 2010-04-21 | | 公開日 | 2010-05-26 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3M7U
 
 | |
3MNX
 
 | | Investigation of global and local effects of radiation damage on porcine pancreatic elastase. Fourth stage of radiation damage | | 分子名称: | Chymotrypsin-like elastase family member 1, SODIUM ION, SULFATE ION | | 著者 | Petrova, T, Ginell, S, Kim, Y, Joachimiak, G, Joachimiak, A. | | 登録日 | 2010-04-22 | | 公開日 | 2010-05-05 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.386 Å) | | 主引用文献 | X-ray-induced deterioration of disulfide bridges at atomic resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4BVW
 
 | | Identification of small molecule inhibitors selective for apo(a) kringles KIV-7, KIV-10 and KV. | | 分子名称: | 1,2,3,4-tetrahydroisoquinoline-6-carboxylic acid, 1,2-ETHANEDIOL, APOLIPOPROTEIN(A), ... | | 著者 | Sandmark, J, Althage, M, Andersson, G.M.K, Antonsson, T, Blaho, S, Bodin, C, Bostrom, J, Chen, Y, Dahlen, A, Eriksson, P.O, Evertsson, E, Fex, T, Fjellstrom, O, Gustafsson, D, Hallberg, C, Hicks, R, Jarkvist, E, Johansson, C, Kalies, I, Kang, D, Svalstedt Karlsson, B, Kartberg, F, Legnehed, A, Lindqvist, A.M, Martinsson, S.A, Moberg, A, Petersson, A.U, Ridderstrom, M, Thelin, A, Tigerstrom, A, Vinblad, J, Xu, B, Knecht, W. | | 登録日 | 2013-06-28 | | 公開日 | 2014-07-16 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Small Molecules Used to Decipher the Pathophysiological Roles of the Kringle Domains Kiv-7, - 10 and Kv of Apolipoprotein(A)
To be Published
|
|
3MHD
 
 | | Crystal structure of DCR3 | | 分子名称: | Tumor necrosis factor receptor superfamily member 6B | | 著者 | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | 登録日 | 2010-04-07 | | 公開日 | 2011-02-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.901 Å) | | 主引用文献 | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
6H3B
 
 | |
