5CLV
 
 | | Crystal Structure of KorA-operator DNA complex (KorA-OA) | | 分子名称: | 5'-D(CP*CP*AP*AP*GP*TP*TP*TP*AP*GP*CP*TP*AP*AP*AP*CP*TP*TP*GP*GP*)-3', TrfB transcriptional repressor protein | | 著者 | White, S.A, Hyde, E.I, Rajasekar, K.V. | | 登録日 | 2015-07-16 | | 公開日 | 2016-04-06 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Flexibility of KorA, a plasmid-encoded, global transcription regulator, in the presence and the absence of its operator.
Nucleic Acids Res., 44, 2016
|
|
4YGI
 
 | |
2BJ1
 
 | |
2BJ9
 
 | | NIKR with bound NICKEL and phosphate | | 分子名称: | NICKEL (II) ION, NICKEL RESPONSIVE REGULATOR, PHOSPHATE ION, ... | | 著者 | Tahirov, T.H, Chivers, P.T. | | 登録日 | 2005-01-31 | | 公開日 | 2005-04-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structure of Pyrococcus Horikoshii Nikr: Nickel Sensing and Implications for the Regulation of DNA Recognition
J.Mol.Biol., 348, 2005
|
|
7JO9
 
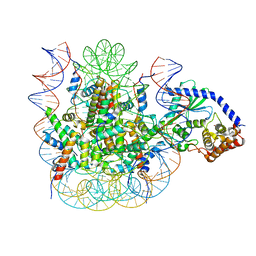 | | 1:1 cGAS-nucleosome complex | | 分子名称: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | 著者 | Boyer, J.A, Spangler, C.J, Strauss, J.D, Cesmat, A.P, Liu, P, McGinty, R.K, Zhang, Q. | | 登録日 | 2020-08-06 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of nucleosome-dependent cGAS inhibition.
Science, 370, 2020
|
|
4TU9
 
 | | STRUCTURE OF U2AF65 VARIANT WITH BRU5G6 DNA | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*DG*U)-3'), GLYCEROL, ... | | 著者 | Jenkins, J.L, McLaughlin, K.J, Agrawal, A.A, Kielkopf, C.L. | | 登録日 | 2014-06-24 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.992 Å) | | 主引用文献 | Structure-guided U2AF65 variant improves recognition and splicing of a defective pre-mRNA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3ULQ
 
 | |
4TU8
 
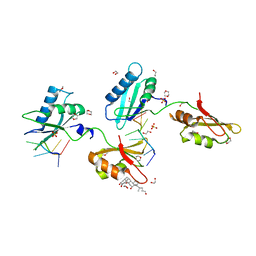 | | STRUCTURE OF U2AF65 VARIANT WITH BRU5A6 DNA | | 分子名称: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*DA*U)-3'), ... | | 著者 | MCLAUGHLIN, K.J, JENKINS, J.L, Agrawal, A.A, KIELKOPF, C.L. | | 登録日 | 2014-06-24 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.918 Å) | | 主引用文献 | Structure-guided U2AF65 variant improves recognition and splicing of a defective pre-mRNA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8SOJ
 
 | |
4TU7
 
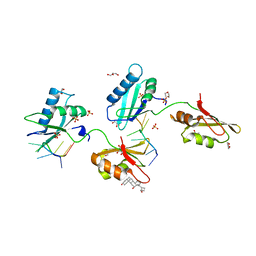 | | Structure of U2AF65 D231V variant with BrU5 DNA | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, DNA (5'-D(*UP*UP*UP*UP*(BRU)P*UP*U)-3'), GLYCEROL, ... | | 著者 | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | 登録日 | 2014-06-24 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.087 Å) | | 主引用文献 | Structure-guided U2AF65 variant improves recognition and splicing of a defective pre-mRNA.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
8XP8
 
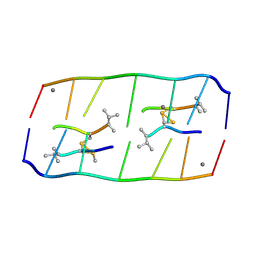 | | Crystal structure of d(ACGmCCGT/ACGGCGT) in complex with Echinomycin | | 分子名称: | 2-CARBOXYQUINOXALINE, DNA (5'-D(P*AP*CP*GP*(5CM)P*CP*GP*T)-3'), DNA (5'-D(P*AP*CP*GP*GP*CP*GP*T)-3'), ... | | 著者 | Hou, M.H, Lin, S.M, Neidle, H. | | 登録日 | 2024-01-03 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-08-28 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | Structural basis of water-mediated cis Watson-Crick/Hoogsteen base-pair formation in non-CpG methylation.
Nucleic Acids Res., 52, 2024
|
|
7JOA
 
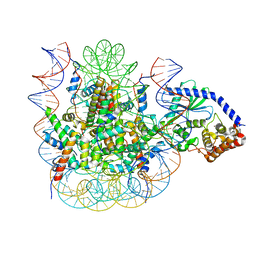 | | 2:1 cGAS-nucleosome complex | | 分子名称: | Cyclic GMP-AMP synthase, DNA (145-MER), Histone H2A type 1, ... | | 著者 | Boyer, J.A, Spangler, C.J, Strauss, J.D, Cesmat, A.P, Liu, P, McGinty, R.K, Zhang, Q. | | 登録日 | 2020-08-06 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of nucleosome-dependent cGAS inhibition.
Science, 370, 2020
|
|
5D1J
 
 | |
1D58
 
 | | THE MOLECULAR STRUCTURE OF A 4'-EPIADRIAMYCIN COMPLEX WITH D(TGATCA) AT 1.7 ANGSTROM RESOLUTION-COMPARISON WITH THE STRUCTURE OF 4'-EPIADRIAMYCIN D(TGTACA) AND D(CGATCG) COMPLEXES | | 分子名称: | 4'-EPIDOXORUBICIN, DNA (5'-D(*TP*GP*AP*TP*CP*A)-3') | | 著者 | Langlois D'Estaintot, B, Gallois, B, Brown, T, Hunter, W.N. | | 登録日 | 1992-02-20 | | 公開日 | 1992-10-15 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | The molecular structure of a 4'-epiadriamycin complex with d(TGATCA) at 1.7A resolution: comparison with the structure of 4'-epiadriamycin d(TGTACA) and d(CGATCG) complexes.
Nucleic Acids Res., 20, 1992
|
|
3TQY
 
 | | Structure of a single-stranded DNA-binding protein (ssb), from Coxiella burnetii | | 分子名称: | Single-stranded DNA-binding protein | | 著者 | Cheung, J, Franklin, M.C, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | 登録日 | 2011-09-09 | | 公開日 | 2011-09-21 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6001 Å) | | 主引用文献 | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
8GLX
 
 | |
7Z7E
 
 | | Crystal structure of p63 DNA binding domain in complex with inhibitory DARPin G4 | | 分子名称: | DARPIN, Isoform 4 of Tumor protein 63, ZINC ION | | 著者 | Strubel, A, Gebel, J, Chaikuad, A, Muenick, P, Doetsch, V. | | 登録日 | 2022-03-15 | | 公開日 | 2022-06-29 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Designed Ankyrin Repeat Proteins as a tool box for analyzing p63.
Cell Death Differ., 29, 2022
|
|
6PUW
 
 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and Bictegravir (BIC) | | 分子名称: | Bictegravir, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | 著者 | Lyumkis, D, Jozwik, I.K, Passos, D. | | 登録日 | 2019-07-18 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6PUZ
 
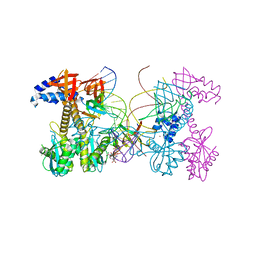 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ446 (compound 4f) | | 分子名称: | 4-azanyl-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-6-[2-(phenylsulfonyl)ethyl]-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | 著者 | Lyumkis, D, Jozwik, I.K, Passos, D. | | 登録日 | 2019-07-18 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6PUY
 
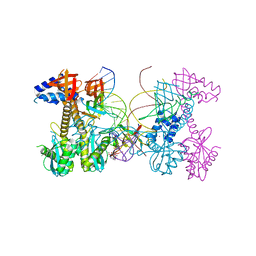 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with magnesium and INSTI XZ426 (compound 4d) | | 分子名称: | 4-amino-N-[(2,4-difluorophenyl)methyl]-1-hydroxy-6-(6-hydroxyhexyl)-2-oxo-1,2-dihydro-1,8-naphthyridine-3-carboxamide, Chimeric Sso7d and HIV-1 integrase, MAGNESIUM ION, ... | | 著者 | Lyumkis, D, Jozwik, I.K, Passos, D. | | 登録日 | 2019-07-18 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
6PUT
 
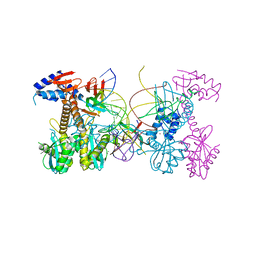 | | Structure of HIV cleaved synaptic complex (CSC) intasome bound with calcium | | 分子名称: | CALCIUM ION, Chimeric Sso7d and HIV-1 integrase, ZINC ION, ... | | 著者 | Lyumkis, D, Jozwik, I.K, Passos, D. | | 登録日 | 2019-07-18 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for strand-transfer inhibitor binding to HIV intasomes.
Science, 367, 2020
|
|
7K9Y
 
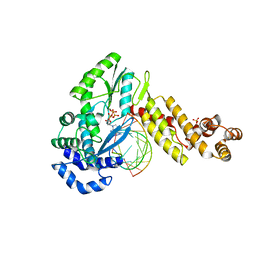 | | GsI-IIC RT Template-Switching Complex (twinned) | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*TP*CP*CP*AP*GP*GP*CP*AP*AP*C)-3'), MAGNESIUM ION, ... | | 著者 | Stamos, J.L, Lentzsch, A.M. | | 登録日 | 2020-09-29 | | 公開日 | 2021-08-18 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural basis for template switching by a group II intron-encoded non-LTR-retroelement reverse transcriptase.
J.Biol.Chem., 297, 2021
|
|
6G0L
 
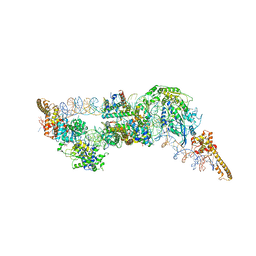 | | Structure of two molecules of the chromatin remodelling enzyme Chd1 bound to a nucleosome | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | 著者 | Sundaramoorthy, R, Owen-hughes, T, Norman, D.G, Hughes, A. | | 登録日 | 2018-03-19 | | 公開日 | 2018-08-22 | | 最終更新日 | 2018-11-21 | | 実験手法 | ELECTRON MICROSCOPY (10 Å) | | 主引用文献 | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome.
Elife, 7, 2018
|
|
7BME
 
 | |
7ET6
 
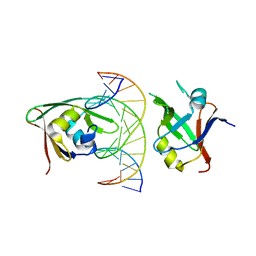 | | Crystal structure of Arabidopsis TEM1 B3-DNA complex | | 分子名称: | AP2/ERF and B3 domain-containing transcription repressor TEM1, FT-RY14-F, FT-RY14-R | | 著者 | Hu, H, Du, J. | | 登録日 | 2021-05-12 | | 公開日 | 2021-09-15 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
