8HXY
 
 | |
5U8S
 
 | | Structure of eukaryotic CMG helicase at a replication fork | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Cell division control protein 45, DNA (26-MER), ... | | 著者 | Li, H, Li, B, Georgescu, R, Yuan, Z, Santos, R, Sun, J, Zhang, D, Yurieva, O, O'Donnell, M.E. | | 登録日 | 2016-12-14 | | 公開日 | 2017-01-25 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (6.1 Å) | | 主引用文献 | Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4LUZ
 
 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | 分子名称: | 5-(4-{[4-(5-carboxyfuran-2-yl)benzyl]oxy}phenyl)-1-(3-methylphenyl)-1H-pyrazole-3-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | 著者 | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | 登録日 | 2013-07-25 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
4LW1
 
 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | 分子名称: | 5-(3-chloro-4-fluorophenyl)furan-2-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | 著者 | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | 登録日 | 2013-07-26 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.631 Å) | | 主引用文献 | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
4LWC
 
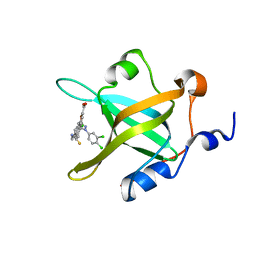 | | Fragment-Based Discovery of a Potent Inhibitor of Replication Protein A Protein-Protein Interactions | | 分子名称: | 5-[3-chloro-4-({4-[1-(3,4-dichlorophenyl)-1H-pyrazol-5-yl]benzyl}carbamothioyl)phenyl]furan-2-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | 著者 | Feldkamp, M.D, Frank, A.O, Kennedy, J.P, Waterson, A.G, Olejnczak, E.O, Pelz, N.F, Patrone, J.D, Vangamudi, B, Camper, D.V, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | 登録日 | 2013-07-26 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.61 Å) | | 主引用文献 | Discovery of a potent inhibitor of replication protein a protein-protein interactions using a fragment-linking approach.
J.Med.Chem., 56, 2013
|
|
8OW1
 
 | | Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome. | | 分子名称: | C0N3, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | 登録日 | 2023-04-26 | | 公開日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
3NH2
 
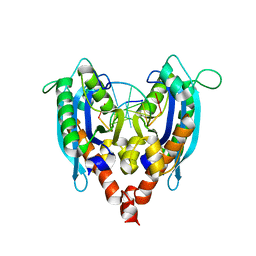 | |
3NH0
 
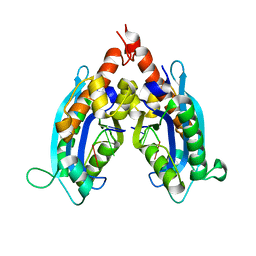 | |
5YUN
 
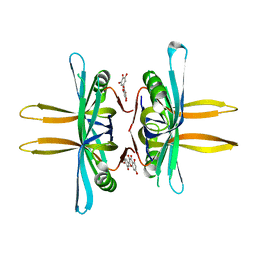 | | Crystal structure of SSB complexed with myc | | 分子名称: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Single-stranded DNA-binding protein | | 著者 | Huang, Y.H, Huang, C.Y. | | 登録日 | 2017-11-22 | | 公開日 | 2018-10-10 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Crystal structure of SSB complexed with inhibitor myricetin.
Biochem. Biophys. Res. Commun., 504, 2018
|
|
3NGZ
 
 | |
3NH1
 
 | |
2AS5
 
 | | Structure of the DNA binding domains of NFAT and FOXP2 bound specifically to DNA. | | 分子名称: | 5'-D(AP*AP*CP*TP*AP*TP*GP*AP*AP*AP*CP*AP*AP*AP*TP*TP*TP*TP*CP*CP*TP*)-3', 5'-D(TP*TP*AP*GP*GP*AP*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*CP*AP*TP*AP*GP*)-3', Forkhead box protein P2, ... | | 著者 | Wu, Y, Stroud, J.C, Borde, M, Bates, D.L, Guo, L, Han, A, Rao, A, Chen, L. | | 登録日 | 2005-08-22 | | 公開日 | 2006-08-08 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | FOXP3 Controls Regulatory T Cell Function through Cooperation with NFAT.
Cell(Cambridge,Mass.), 126, 2006
|
|
4L9J
 
 | |
5FUR
 
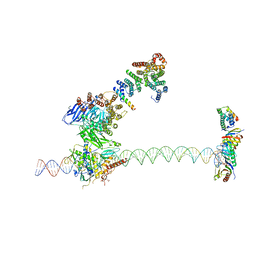 | | Structure of human TFIID-IIA bound to core promoter DNA | | 分子名称: | SUPER CORE PROMOTER, TATA-BOX-BINDING PROTEIN, TRANSCRIPTION INITIATION FACTOR IIA SUBUNIT 1, ... | | 著者 | Louder, R.K, He, Y, Lopez-Blanco, J.R, Fang, J, Chacon, P, Nogales, E. | | 登録日 | 2016-01-29 | | 公開日 | 2016-04-06 | | 最終更新日 | 2017-08-02 | | 実験手法 | ELECTRON MICROSCOPY (8.5 Å) | | 主引用文献 | Structure of Promoter-Bound TFIID and Model of Human Pre-Initiation Complex Assembly.
Nature, 531, 2016
|
|
6V92
 
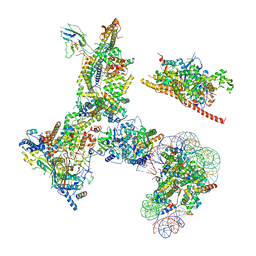 | | RSC-NCP | | 分子名称: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC14, ... | | 著者 | Patel, A.B, Moore, C.M, Greber, B.J, Nogales, E. | | 登録日 | 2019-12-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (20 Å) | | 主引用文献 | Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement.
Elife, 8, 2019
|
|
7PDS
 
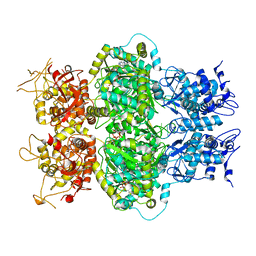 | | The structure of PriRep1 with dsDNA | | 分子名称: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Similar to D. nodosus vapE, ... | | 著者 | Qiao, C.C, Mir Sanchis, I. | | 登録日 | 2021-08-06 | | 公開日 | 2022-07-13 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Staphylococcal self-loading helicases couple the staircase mechanism with inter domain high flexibility.
Nucleic Acids Res., 50, 2022
|
|
2HZD
 
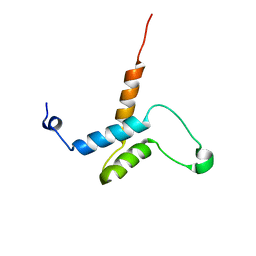 | |
5U8T
 
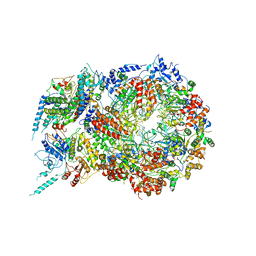 | | Structure of Eukaryotic CMG Helicase at a Replication Fork and Implications | | 分子名称: | Cell division control protein 45, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA replication complex GINS protein PSF1, ... | | 著者 | Li, B, Georgescu, R, Yuan, Z, Santos, R, Sun, J, Zhang, D, Yurieva, O, Li, H, O'Donnell, M.E. | | 登録日 | 2016-12-15 | | 公開日 | 2017-02-08 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6BOV
 
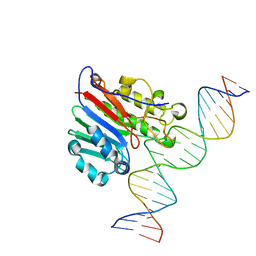 | |
6BOQ
 
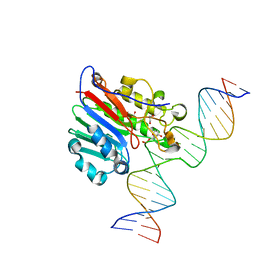 | | Human APE1 substrate complex with an A/A mismatch adjacent the THF | | 分子名称: | 1,2-ETHANEDIOL, 21-mer DNA, DNA-(apurinic or apyrimidinic site) lyase | | 著者 | Freudenthal, B.D, Whitaker, A.M, Fairlamb, M.S. | | 登録日 | 2017-11-20 | | 公開日 | 2018-08-15 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Apurinic/apyrimidinic (AP) endonuclease 1 processing of AP sites with 5' mismatches.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6BOR
 
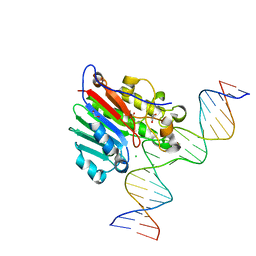 | | Human APE1 substrate complex with an G/G mismatch adjacent the THF | | 分子名称: | 1,2-ETHANEDIOL, 21-mer DNA, CHLORIDE ION, ... | | 著者 | Freudenthal, B.D, Whitaker, A.M, Fairlamb, M.S. | | 登録日 | 2017-11-20 | | 公開日 | 2018-08-15 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Apurinic/apyrimidinic (AP) endonuclease 1 processing of AP sites with 5' mismatches.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6BOT
 
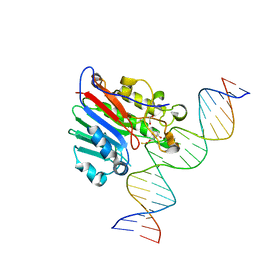 | | Human APE1 substrate complex with an C/C mismatch adjacent the THF | | 分子名称: | 1,2-ETHANEDIOL, 21-mer DNA, DNA-(apurinic or apyrimidinic site) lyase | | 著者 | Freudenthal, B.D, Whitaker, A.M, Fairlamb, M.S. | | 登録日 | 2017-11-20 | | 公開日 | 2018-08-15 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Apurinic/apyrimidinic (AP) endonuclease 1 processing of AP sites with 5' mismatches.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
6BOW
 
 | |
6BOS
 
 | |
6BOU
 
 | |
