1R9T
 
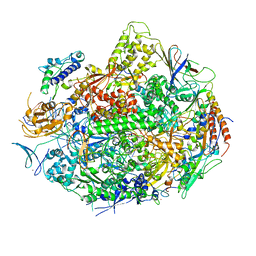 | | RNA POLYMERASE II STRAND SEPARATED ELONGATION COMPLEX, MISMATCHED NUCLEOTIDE | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, DNA nontemplate strand, DNA template strand, ... | | 著者 | Westover, K.D, Bushnell, D.A, Kornberg, R.D. | | 登録日 | 2003-10-30 | | 公開日 | 2004-11-16 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center.
Cell(Cambridge,Mass.), 119, 2004
|
|
2E42
 
 | | Crystal structure of C/EBPbeta Bzip homodimer V285A mutant bound to A High Affinity DNA fragment | | 分子名称: | CCAAT/enhancer-binding protein beta, DNA (5'-D(P*DAP*DAP*DTP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DCP*DCP*DT)-3'), DNA (5'-D(P*DTP*DAP*DGP*DGP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DAP*DT)-3') | | 著者 | Tahirov, T.H, Inoue-Bungo, T, Sato, K, Shiina, M, Hamada, K, Ogata, K. | | 登録日 | 2006-12-01 | | 公開日 | 2007-12-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural Basis for Flexible Base Recognition by C/Ebpbeta
To be Published
|
|
2E43
 
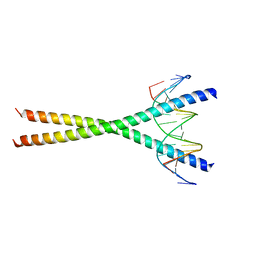 | | Crystal structure of C/EBPbeta Bzip homodimer K269A mutant bound to A High Affinity DNA fragment | | 分子名称: | CCAAT/enhancer-binding protein beta, DNA (5'-D(P*DAP*DAP*DTP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DCP*DCP*DT)-3'), DNA (5'-D(P*DTP*DAP*DGP*DGP*DAP*DTP*DTP*DGP*DCP*DGP*DCP*DAP*DAP*DTP*DAP*DT)-3') | | 著者 | Tahirov, T.H, Inoue-Bungo, T, Sato, K, Shiina, M, Hamada, K, Ogata, K. | | 登録日 | 2006-12-01 | | 公開日 | 2007-12-18 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural Basis for Flexible Base Recognition by C/Ebpbeta
To be Published
|
|
4PXI
 
 | | Elucidation of the Structural and Functional Mechanism of Action of the TetR Family Protein, CprB from S. coelicolor A3(2) | | 分子名称: | CprB, DNA (5'-D(*AP*CP*AP*TP*AP*CP*GP*GP*GP*AP*CP*GP*CP*CP*CP*CP*GP*TP*TP*TP*AP*T)-3'), DNA (5'-D(*AP*TP*AP*AP*AP*CP*GP*GP*GP*GP*CP*GP*TP*CP*CP*CP*GP*TP*AP*TP*GP*T)-3') | | 著者 | Hussain, B, Ruchika, B, Aruna, B, Ruchi, A. | | 登録日 | 2014-03-24 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural and functional basis of transcriptional regulation by TetR family protein CprB from S. coelicolor A3(2)
Nucleic Acids Res., 42, 2014
|
|
5U8T
 
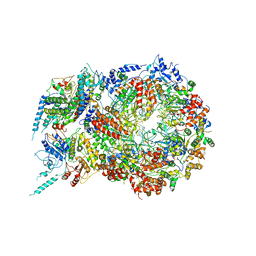 | | Structure of Eukaryotic CMG Helicase at a Replication Fork and Implications | | 分子名称: | Cell division control protein 45, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA replication complex GINS protein PSF1, ... | | 著者 | Li, B, Georgescu, R, Yuan, Z, Santos, R, Sun, J, Zhang, D, Yurieva, O, Li, H, O'Donnell, M.E. | | 登録日 | 2016-12-15 | | 公開日 | 2017-02-08 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (4.9 Å) | | 主引用文献 | Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5W0R
 
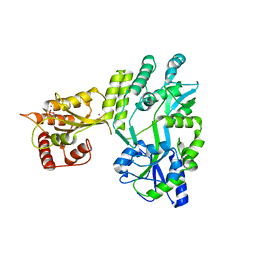 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with cacodylic acid | | 分子名称: | CACODYLATE ION, CALCIUM ION, MBP fused activation-induced cytidine deaminase, ... | | 著者 | Qiao, Q, Wang, L, Wu, H. | | 登録日 | 2017-05-31 | | 公開日 | 2017-08-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
4URM
 
 | | Crystal Structure of Staph GyraseB 24kDa in complex with Kibdelomycin | | 分子名称: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA GYRASE SUBUNIT B | | 著者 | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | 登録日 | 2014-06-30 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
5N8R
 
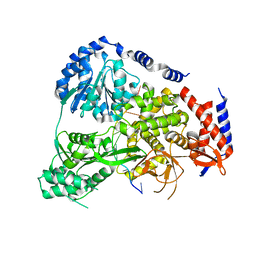 | | Crystal Structure of Drosophilia DHX36 helicase in complex with GAGCACTGC | | 分子名称: | CG9323, isoform A, DNA (5'-D(P*GP*AP*GP*CP*AP*CP*TP*GP*C)-3') | | 著者 | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | 登録日 | 2017-02-24 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
4X4I
 
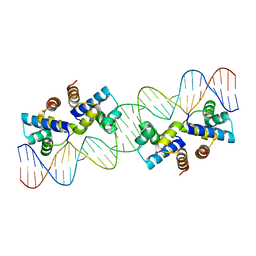 | |
4X4H
 
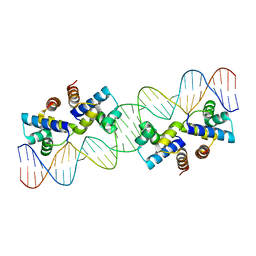 | |
1HXD
 
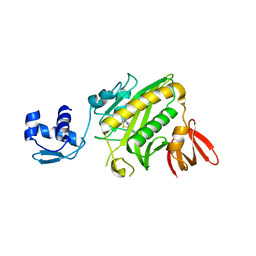 | | CRYSTAL STRUCTURE OF E. COLI BIOTIN REPRESSOR WITH BOUND BIOTIN | | 分子名称: | BIOTIN, BIRA BIFUNCTIONAL PROTEIN | | 著者 | Kwon, K, Streaker, E.D, Ruparelia, S, Beckett, D. | | 登録日 | 2001-01-12 | | 公開日 | 2001-05-30 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Corepressor-induced organization and assembly of the biotin repressor: a model for allosteric activation of a transcriptional regulator.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
7S8D
 
 | | Structure of DNA-free SgrAI | | 分子名称: | CALCIUM ION, SgraIR restriction enzyme | | 著者 | Horton, N.C. | | 登録日 | 2021-09-17 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Pretransition state and apo structures of the filament-forming enzyme SgrAI elucidate mechanisms of activation and substrate specificity.
J.Biol.Chem., 298, 2022
|
|
5N98
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with TAGGGTTTT | | 分子名称: | CG9323, isoform A, DNA (5'-D(P*TP*AP*GP*GP*GP*TP*TP*TP*T)-3'), ... | | 著者 | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | 登録日 | 2017-02-24 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.756 Å) | | 主引用文献 | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N9D
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with GGGTTAGGGT | | 分子名称: | CG9323, isoform A, DNA (5'-D(P*GP*GP*GP*TP*TP*AP*GP*GP*GP*T)-3') | | 著者 | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | 登録日 | 2017-02-24 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N90
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with TTGTGGTGT | | 分子名称: | CG9323, isoform A, DNA (5'-D(P*TP*TP*GP*TP*GP*GP*TP*GP*T)-3'), ... | | 著者 | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | 登録日 | 2017-02-24 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.069 Å) | | 主引用文献 | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N8S
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with polyT | | 分子名称: | CG9323, isoform A, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | 著者 | Chen, W.-F, Rety, S, Hai-Lei Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | 登録日 | 2017-02-24 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.88 Å) | | 主引用文献 | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
4X4G
 
 | |
5N96
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with AGGGTTTTTT | | 分子名称: | CG9323, isoform A, DNA (5'-D(P*AP*GP*GP*GP*TP*TP*TP*TP*TP*T)-3'), ... | | 著者 | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | 登録日 | 2017-02-24 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.716 Å) | | 主引用文献 | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
5N9A
 
 | | Crystal Structure of Drosophila DHX36 helicase in complex with GTTAGGGTT | | 分子名称: | CG9323, isoform A, DNA (5'-D(P*GP*TP*TP*AP*GP*GP*GP*TP*T)-3') | | 著者 | Chen, W.-F, Rety, S, Guo, H.-L, Wu, W.-Q, Liu, N.-N, Liu, Q.-W, Dai, Y.-X, Xi, X.-G. | | 登録日 | 2017-02-24 | | 公開日 | 2018-03-14 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.036 Å) | | 主引用文献 | Molecular Mechanistic Insights into Drosophila DHX36-Mediated G-Quadruplex Unfolding: A Structure-Based Model.
Structure, 26, 2018
|
|
8OW1
 
 | | Cryo-EM structure of the yeast Inner kinetochore bound to a CENP-A nucleosome. | | 分子名称: | C0N3, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Dendooven, T.D, Zhang, Z, Yang, J, McLaughlin, S, Schwabb, J, Scheres, S, Yatskevich, S, Barford, D. | | 登録日 | 2023-04-26 | | 公開日 | 2023-08-09 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Cryo-EM structure of the complete inner kinetochore of the budding yeast point centromere.
Sci Adv, 9, 2023
|
|
6LND
 
 | |
4X4F
 
 | |
8HY0
 
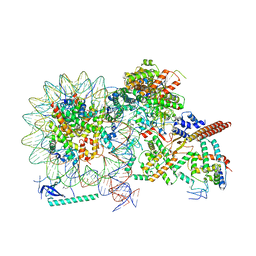 | |
8HXY
 
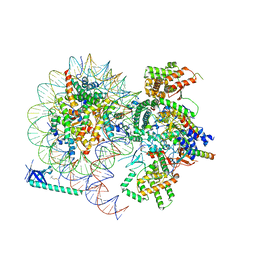 | |
8TOF
 
 | | Rpd3S bound to an H3K36Cme3 modified nucleosome | | 分子名称: | Chromatin modification-related protein EAF3, DNA (176-MER), Histone H2A, ... | | 著者 | Markert, J.W, Vos, S.M, Farnung, L. | | 登録日 | 2023-08-03 | | 公開日 | 2024-01-17 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Structure of the complete Saccharomyces cerevisiae Rpd3S-nucleosome complex.
Nat Commun, 14, 2023
|
|
