1QQG
 
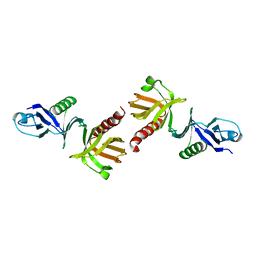 | |
4BE1
 
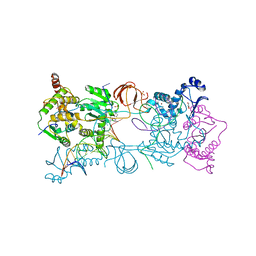 | | PFV intasome with inhibitor XZ-116 | | 分子名称: | 17 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (TRANSFERRED STRAND), 19 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (NON-TRANSFERRED STRAND), 2-(3-chloro-2-fluorobenzyl)-6,7-dihydroxy-2,3-dihydro-1H-isoindol-1-one, ... | | 著者 | Hare, S, Cherepanov, P. | | 登録日 | 2012-10-08 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | Activities, Crystal Structures and Molecular Dynamics of Dihydro-1H-Isoindole Derivatives, Inhibitors of HIV-1 Integrase.
Acs Chem.Biol., 8, 2013
|
|
3BD6
 
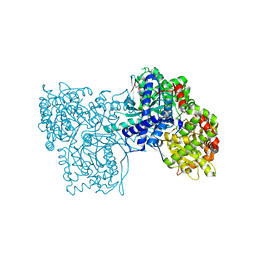 | | Glycogen Phosphorylase complex with 1(-D-ribofuranosyl) cyanuric acid | | 分子名称: | 1-beta-D-ribofuranosyl-1,3,5-triazinane-2,4,6-trione, Glycogen phosphorylase, muscle form | | 著者 | Sovantzis, D.A, Hadjiloi, T, Hayes, J.M, Zographos, S.E, Chrysina, E.D, Oikonomakos, N.G. | | 登録日 | 2007-11-14 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | D-Glucopyranosyl pyrimidine nucleoside binding to muscle glycogen phosphorylase b
To be Published
|
|
5W58
 
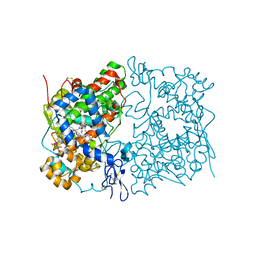 | | Crystal Complex of Cyclooxygenase-2: (S)-ARN-2508 (a dual COX and FAAH inhibitor) | | 分子名称: | (2S)-2-{2-fluoro-3'-[(hexylcarbamoyl)oxy][1,1'-biphenyl]-4-yl}propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Xu, S, Goodman, M.C, Banerjee, S, Piomelli, D, Marnett, L.J. | | 登録日 | 2017-06-14 | | 公開日 | 2018-01-31 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.267 Å) | | 主引用文献 | Dual cyclooxygenase-fatty acid amide hydrolase inhibitor exploits novel binding interactions in the cyclooxygenase active site.
J. Biol. Chem., 293, 2018
|
|
3KTD
 
 | |
4HBQ
 
 | |
2P4U
 
 | | Crystal structure of acid phosphatase 1 (Acp1) from Mus musculus | | 分子名称: | Acid phosphatase 1, PHOSPHATE ION | | 著者 | Bonanno, J.B, Freeman, J, Bain, K.T, Wu, B, Xu, W, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2007-03-13 | | 公開日 | 2007-03-20 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
4HEE
 
 | | Crystal structure of PPARgamma in complex with compound 13 | | 分子名称: | 5-benzyl-2-ethyl-3-{(1S)-5-[2-(1H-tetrazol-5-yl)phenyl]-2,3-dihydro-1H-inden-1-yl}-3,5-dihydro-4H-imidazo[4,5-c]pyridin-4-one, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | 著者 | Han, S. | | 登録日 | 2012-10-03 | | 公開日 | 2013-08-21 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Design, synthesis, and evaluation of imidazo[4,5-c]pyridin-4-one derivatives with dual activity at angiotensin II type 1 receptor and peroxisome proliferator-activated receptor-gamma
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4BE2
 
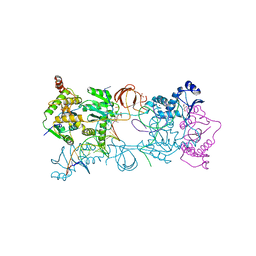 | | PFV intasome with inhibitor XZ-259 | | 分子名称: | 17 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (TRANSFERRED STRAND) *AP*CP*A)-3', 19 NUCLEOTIDE PREPROCESSED PFV DONOR DNA (NON-TRANSFERRED STRAND), 2-(3-chloro-4-fluorobenzyl)-6,7-dihydroxy-N,N-dimethyl-1-oxo-2,3-dihydro-1H-isoindole-4-sulfonamide, ... | | 著者 | Hare, S, Cherepanov, P. | | 登録日 | 2012-10-08 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.38 Å) | | 主引用文献 | Activities, Crystal Structures and Molecular Dynamics of Dihydro-1H-Isoindole Derivatives, Inhibitors of HIV-1 Integrase.
Acs Chem.Biol., 8, 2013
|
|
2AAA
 
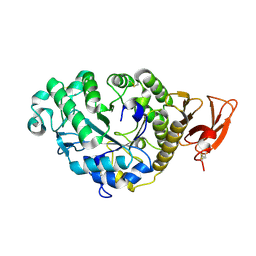 | | CALCIUM BINDING IN ALPHA-AMYLASES: AN X-RAY DIFFRACTION STUDY AT 2.1 ANGSTROMS RESOLUTION OF TWO ENZYMES FROM ASPERGILLUS | | 分子名称: | ALPHA-AMYLASE, CALCIUM ION | | 著者 | Brady, L, Brzozowski, A.M, Derewenda, Z, Dodson, E.J, Dodson, G.G. | | 登録日 | 1991-02-27 | | 公開日 | 1993-07-15 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Calcium binding in alpha-amylases: an X-ray diffraction study at 2.1-A resolution of two enzymes from Aspergillus.
Biochemistry, 29, 1990
|
|
3WF5
 
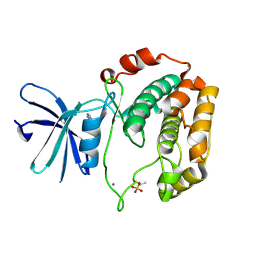 | | Crystal structure of S6K1 kinase domain in complex with a pyrazolopyrimidine derivative 4-[4-(1H-benzimidazol-2-yl)piperidin-1-yl]-1H-pyrazolo[3,4-d]pyrimidine | | 分子名称: | 4-[4-(1H-benzimidazol-2-yl)piperidin-1-yl]-1H-pyrazolo[3,4-d]pyrimidine, Ribosomal protein S6 kinase beta-1, ZINC ION | | 著者 | Niwa, H, Shirouzu, M, Yokoyama, S. | | 登録日 | 2013-07-17 | | 公開日 | 2014-08-06 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Crystal structures of the S6K1 kinase domain in complexes with inhibitors
J.Struct.Funct.Genom., 15, 2014
|
|
3WPY
 
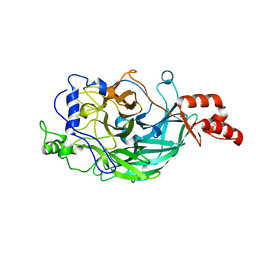 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase mutant T47S/S200T/F447V/P500S | | 分子名称: | Beta-fructofuranosidase | | 著者 | Yokoi, G, Mori, M, Sato, S, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | 登録日 | 2014-01-17 | | 公開日 | 2014-03-12 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Enhancing thermostability and the structural characterization of Microbacterium saccharophilum K-1 beta-fructofuranosidase
Appl.Microbiol.Biotechnol., 98, 2014
|
|
5OI3
 
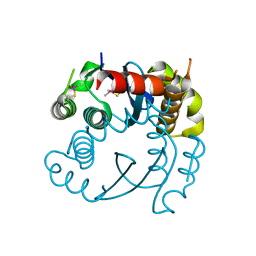 | |
3J3Q
 
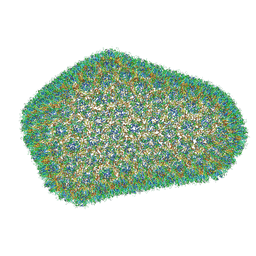 | |
1VSB
 
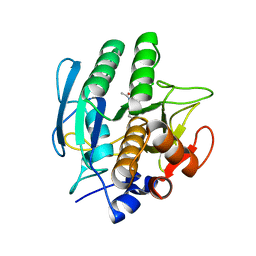 | | SUBTILISIN CARLSBERG L-PARA-CHLOROPHENYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | 分子名称: | SUBTILISIN CARLSBERG, TYPE VIII | | 著者 | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | 登録日 | 1997-09-17 | | 公開日 | 1998-03-18 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
5OH0
 
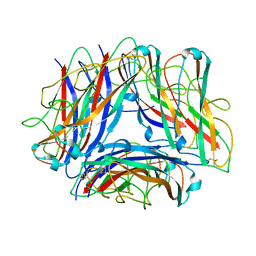 | | The Cryo-Electron Microscopy Structure of the Type 1 Chaperone-Usher Pilus Rod | | 分子名称: | Type-1 fimbrial protein, A chain | | 著者 | Hospenthal, M.K, Costa, T.R.D, Redzej, A, Waksman, G. | | 登録日 | 2017-07-13 | | 公開日 | 2017-11-22 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | The Cryoelectron Microscopy Structure of the Type 1 Chaperone-Usher Pilus Rod.
Structure, 25, 2017
|
|
4JPE
 
 | | Spirocyclic Beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 (BACE1) Inhibitors | | 分子名称: | (4R)-2-amino-1,3',3'-trimethyl-7'-(pyrimidin-5-yl)-3',4'-dihydro-2'H-spiro[imidazole-4,1'-naphthalen]-5(1H)-one, Beta-secretase 1, NICKEL (II) ION | | 著者 | Vigers, G.P.A, Smith, D. | | 登録日 | 2013-03-19 | | 公開日 | 2013-04-10 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Spirocyclic beta-site amyloid precursor protein cleaving enzyme 1 (BACE1) inhibitors: from hit to lowering of cerebrospinal fluid (CSF) amyloid beta in a higher species.
J.Med.Chem., 56, 2013
|
|
5OEZ
 
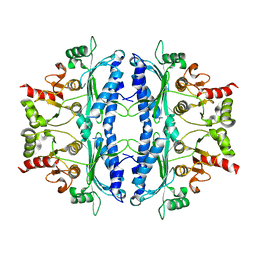 | | Crystal structure of Leishmania major fructose-1,6-bisphosphatase in apo form. | | 分子名称: | FBP protein | | 著者 | Yuan, M, Vasquez-Valdivieso, M.G, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | 登録日 | 2017-07-10 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Structures of Leishmania Fructose-1,6-Bisphosphatase Reveal Species-Specific Differences in the Mechanism of Allosteric Inhibition.
J. Mol. Biol., 429, 2017
|
|
7Z6H
 
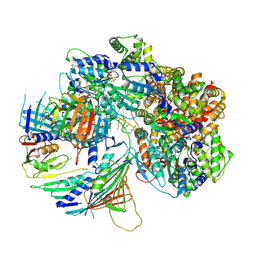 | | Structure of DNA-bound human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp | | 分子名称: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1,Cell cycle checkpoint protein RAD17, Checkpoint protein HUS1, ... | | 著者 | Day, M, Oliver, A.W, Pearl, L.H. | | 登録日 | 2022-03-11 | | 公開日 | 2022-05-04 | | 最終更新日 | 2025-10-01 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | Structure of the human RAD17-RFC clamp loader and 9-1-1 checkpoint clamp bound to a dsDNA-ssDNA junction.
Nucleic Acids Res., 50, 2022
|
|
7JYE
 
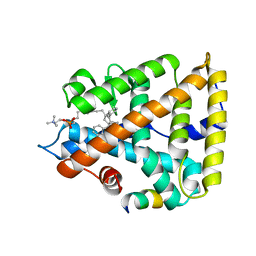 | | Human Liver Receptor Homolog-1 in Complex with 9ChoP and a Fragment of Tif2 | | 分子名称: | 9-[(3~{a}~{R},6~{R},6~{a}~{R})-6-oxidanyl-3-phenyl-3~{a}-(1-phenylethenyl)-4,5,6,6~{a}-tetrahydro-1~{H}-pentalen-2-yl]nonyl 2-(trimethyl-$l^{4}-azanyl)ethyl hydrogen phosphate, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | 著者 | D'Agostino, E.H, Mays, S.G, Ortlund, E.A. | | 登録日 | 2020-08-30 | | 公開日 | 2022-03-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.551 Å) | | 主引用文献 | Tapping into a phospholipid-LRH-1 axis yields a powerful anti-inflammatory agent with in vivo activity against colitis
To Be Published
|
|
3RU5
 
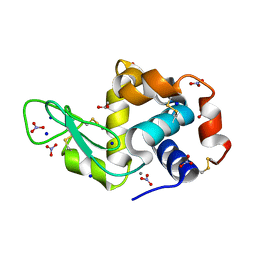 | | Silver Metallated Hen Egg White Lysozyme at 1.35 A | | 分子名称: | 1,2-ETHANEDIOL, Lysozyme C, NITRATE ION, ... | | 著者 | Leeper, T.C, Panzner, M.J, Bilinovich, S.M. | | 登録日 | 2011-05-04 | | 公開日 | 2011-11-23 | | 最終更新日 | 2024-11-27 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Silver metallation of hen egg white lysozyme: X-ray crystal structure and NMR studies.
Chem.Commun.(Camb.), 47, 2011
|
|
1YWK
 
 | |
3WPZ
 
 | | Microbacterium saccharophilum K-1 beta-fructofuranosidase mutant T47S/S200T/F447P/F470Y/P500S | | 分子名称: | Beta-fructofuranosidase | | 著者 | Yokoi, G, Mori, M, Sato, S, Miyazaki, T, Nishikawa, A, Tonozuka, T. | | 登録日 | 2014-01-17 | | 公開日 | 2014-03-12 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Enhancing thermostability and the structural characterization of Microbacterium saccharophilum K-1 beta-fructofuranosidase
Appl.Microbiol.Biotechnol., 98, 2014
|
|
4GNV
 
 | |
5ELV
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L504-N775) in complex with glutamate and BPAM-521 at 1.92 A resolution | | 分子名称: | 4-Cyclopropyl-3,4-dihydro-7-hydroxy-2H-1,2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, CHLORIDE ION, ... | | 著者 | Krintel, C, Juknaite, L, Frydenvang, K, Kastrup, J.S. | | 登録日 | 2015-11-05 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Enthalpy-Entropy Compensation in the Binding of Modulators at Ionotropic Glutamate Receptor GluA2.
Biophys.J., 110, 2016
|
|
