7L28
 
 | | Crystal structure of the catalytic domain of human PDE3A bound to Trequinsin | | 分子名称: | (2E)-9,10-dimethoxy-3-methyl-2-[(2,4,6-trimethylphenyl)imino]-2,3,6,7-tetrahydro-4H-pyrimido[6,1-a]isoquinolin-4-one, ACETATE ION, MAGNESIUM ION, ... | | 著者 | Horner, S.W, Garvie, C. | | 登録日 | 2020-12-16 | | 公開日 | 2021-06-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7L27
 
 | |
7L29
 
 | |
7KWE
 
 | |
6LF9
 
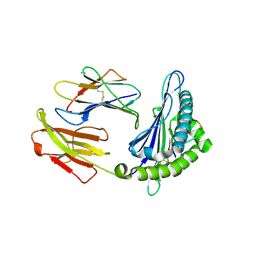 | | Crystal structure of pSLA-1*1301 complex with dodecapeptide RVEDVTNTAEYW | | 分子名称: | ARG-VAL-GLU-ASP-VAL-THR-ASN-THR-ALA-GLU-TYR-TRP, Beta-2-microglobulin, MHC class I antigen | | 著者 | Wei, X.H, Wang, S, Zhang, N.Z, Xia, C. | | 登録日 | 2019-11-30 | | 公開日 | 2021-03-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Peptidomes and Structures Illustrate How SLA-I Micropolymorphism Influences the Preference of Binding Peptide Length.
Front Immunol, 2022
|
|
6YSA
 
 | | Crystal structure of Arabidopsis thaliana legumain isoform beta in zymogen state | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, SULFATE ION, ... | | 著者 | Dall, E, Zauner, F.B, Brandstetter, H. | | 登録日 | 2020-04-21 | | 公開日 | 2020-07-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.01 Å) | | 主引用文献 | Structural and functional studies ofArabidopsis thalianalegumain beta reveal isoform specific mechanisms of activation and substrate recognition.
J.Biol.Chem., 295, 2020
|
|
7K4F
 
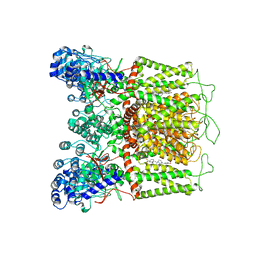 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor 31 | | 分子名称: | 5-[(4-{cis-4-[3-(trifluoromethyl)phenyl]cyclohexyl}piperazin-1-yl)methyl]pyridin-2(1H)-one, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | 著者 | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | 登録日 | 2020-09-15 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.75 Å) | | 主引用文献 | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7K4D
 
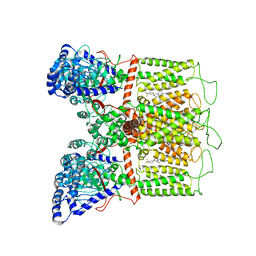 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor 3OG | | 分子名称: | 5-[(4-{trans-4-hydroxy-4-[3-(trifluoromethyl)phenyl]cyclohexyl}piperazin-1-yl)methyl]pyridin-2(1H)-one, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | 著者 | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | 登録日 | 2020-09-15 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7K4B
 
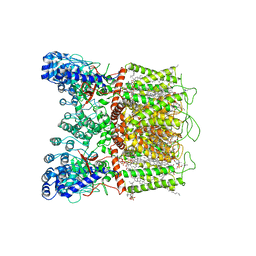 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor cis-22a | | 分子名称: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[cis-4-(3-methylphenyl)cyclohexyl]-4-(pyridin-3-yl)piperazine, CALCIUM ION, ... | | 著者 | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | 登録日 | 2020-09-15 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
1MZA
 
 | | crystal structure of human pro-granzyme K | | 分子名称: | pro-granzyme K | | 著者 | Hink-Schauer, C, Estebanez-Perpina, E, Wilharm, E, Fuentes-Prior, P, Klinkert, W, Bode, W, Jenne, D.E. | | 登録日 | 2002-10-07 | | 公開日 | 2003-01-14 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | The 2.2-A Crystal Structure of Human Pro-granzyme K Reveals a Rigid Zymogen with Unusual Features
J.BIOL.CHEM., 277, 2002
|
|
7JTW
 
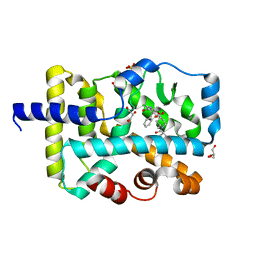 | | Crystal structure of RORgt with compound (4R)-6-[(2,5-dichloro-3-{[(2R,4R)-1-(cyclopentanecarbonyl)-2-methylpiperidin-4-yl]oxy}phenyl)amino]-6-oxo-4-phenylhexanoic acid | | 分子名称: | (4R)-6-[(2,5-dichloro-3-{[(2R,4R)-1-(cyclopentanecarbonyl)-2-methylpiperidin-4-yl]oxy}phenyl)amino]-6-oxo-4-phenylhexanoic acid, GLYCEROL, RAR-related orphan receptor C isoform a variant, ... | | 著者 | Min, X, Wang, Z. | | 登録日 | 2020-08-18 | | 公開日 | 2021-02-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery of 6-Oxo-4-phenyl-hexanoic acid derivatives as ROR gamma t inverse agonists showing favorable ADME profile.
Bioorg.Med.Chem.Lett., 36, 2021
|
|
7K4C
 
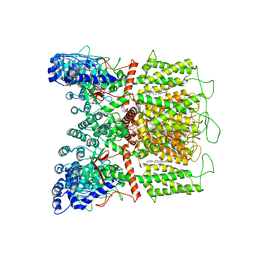 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor Br-cis-22a | | 分子名称: | 1-(5-bromopyridin-3-yl)-4-[cis-4-(3-methylphenyl)cyclohexyl]piperazine, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | 著者 | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | 登録日 | 2020-09-15 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.78 Å) | | 主引用文献 | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
4QRG
 
 | |
4QRV
 
 | |
6MGK
 
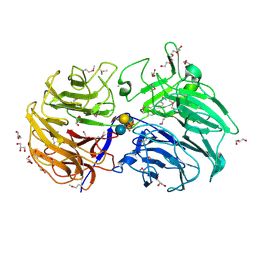 | | Crystal structure of the catalytic domain from GH74 enzyme PoGH74 from Paenibacillus odorifer, in complex with XLX xyloglucan | | 分子名称: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, GLYCEROL, ... | | 著者 | Stogios, P.J, Skarina, T, Nocek, B, Arnal, G, Brumer, H, Savchenko, A. | | 登録日 | 2018-09-14 | | 公開日 | 2019-01-23 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural enzymology reveals the molecular basis of substrate regiospecificity and processivity of an exemplar bacterial glycoside hydrolase family 74endo-xyloglucanase.
Biochem. J., 475, 2018
|
|
7LRD
 
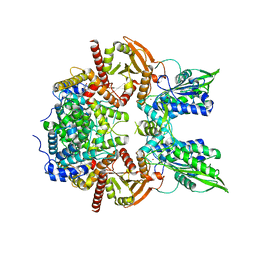 | | Cryo-EM of the SLFN12-PDE3A complex: Consensus subset model | | 分子名称: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, MANGANESE (II) ION, ... | | 著者 | Fuller, J.R, Garvie, C.W, Lemke, C.T. | | 登録日 | 2021-02-16 | | 公開日 | 2021-06-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.22 Å) | | 主引用文献 | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7LRE
 
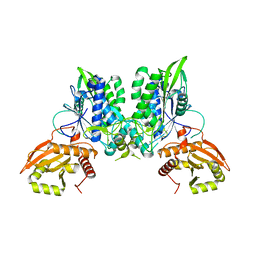 | |
7LRC
 
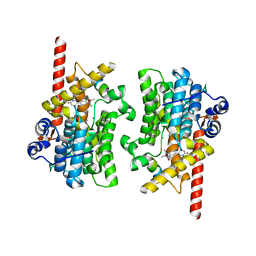 | | Cryo-EM of the SLFN12-PDE3A complex: PDE3A body refinement | | 分子名称: | (4~{R})-3-[4-(diethylamino)-3-[oxidanyl(oxidanylidene)-$l^{4}-azanyl]phenyl]-4-methyl-4,5-dihydro-1~{H}-pyridazin-6-one, MAGNESIUM ION, MANGANESE (II) ION, ... | | 著者 | Fuller, J.R, Garvie, C.W, Lemke, C.T. | | 登録日 | 2021-02-16 | | 公開日 | 2021-06-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (2.97 Å) | | 主引用文献 | Structure of PDE3A-SLFN12 complex reveals requirements for activation of SLFN12 RNase.
Nat Commun, 12, 2021
|
|
7JZI
 
 | |
7JZ4
 
 | |
7JZK
 
 | |
7JZ1
 
 | |
6XRT
 
 | |
7K4E
 
 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor 30 | | 分子名称: | 5-({4-[(1R,4S)-3'-methyl[1,2,3,4-tetrahydro[1,1'-biphenyl]]-4-yl]piperazin-1-yl}methyl)pyridin-2(1H)-one, Transient receptor potential cation channel subfamily V member 6 | | 著者 | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | 登録日 | 2020-09-15 | | 公開日 | 2020-12-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (4.34 Å) | | 主引用文献 | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7LNA
 
 | | Infectious mammalian prion fibril (263K scrapie) | | 分子名称: | Major prion protein | | 著者 | Kraus, A, Hoyt, F, Schwartz, C.L, Hansen, B, Hughson, A.G, Artikis, E, Race, B, Caughey, B. | | 登録日 | 2021-02-06 | | 公開日 | 2021-09-01 | | 最終更新日 | 2021-11-17 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | High-resolution structure and strain comparison of infectious mammalian prions.
Mol.Cell, 81, 2021
|
|
