2MKO
 
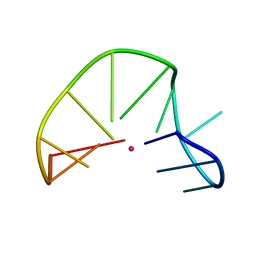 | | G-triplex structure and formation propensity | | 分子名称: | DNA_(5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*G)-3'), POTASSIUM ION | | 著者 | Cerofolini, L, Fragai, M, Giachetti, A, Limongelli, V, Luchinat, C, Novellino, E, Parrinello, M, Randazzo, A. | | 登録日 | 2014-02-11 | | 公開日 | 2014-11-19 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | G-triplex structure and formation propensity.
Nucleic Acids Res., 42, 2014
|
|
2M27
 
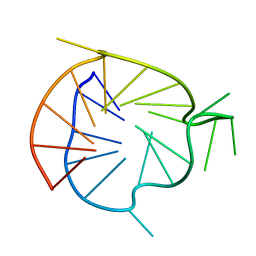 | | Major G-quadruplex structure formed in human VEGF promoter, a monomeric parallel-stranded quadruplex | | 分子名称: | DNA_(5'-D(*CP*GP*GP*GP*GP*CP*GP*GP*GP*CP*CP*TP*TP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3')_ | | 著者 | Agrawal, P, Hatzakis, E, Guo, K, Carver, M, Yang, D. | | 登録日 | 2012-12-14 | | 公開日 | 2013-09-18 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the major G-quadruplex formed in the human VEGF promoter in K+: insights into loop interactions of the parallel G-quadruplexes.
Nucleic Acids Res., 41, 2013
|
|
8JL0
 
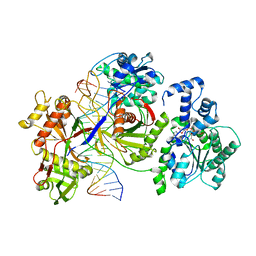 | | Cryo-EM structure of the prokaryotic SPARSA system complex | | 分子名称: | DNA (5'-D(P*AP*CP*GP*AP*CP*GP*TP*CP*TP*AP*AP*GP*AP*AP*AP*CP*CP*AP*TP*TP*AP*T)-3'), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Piwi domain protein, ... | | 著者 | Xu, X, Zhen, X, Long, F. | | 登録日 | 2023-06-02 | | 公開日 | 2024-01-24 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structural basis of antiphage immunity generated by a prokaryotic Argonaute-associated SPARSA system.
Nat Commun, 15, 2024
|
|
4BHC
 
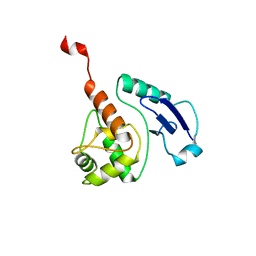 | | CRYSTAL STRUCTURE OF THE M. TUBERCULOSIS O6-METHYLGUANINE METHYLTRANSFERASE R37L VARIANT | | 分子名称: | METHYLATED-DNA--PROTEIN-CYSTEINE METHYLTRANSFERASE | | 著者 | Miggiano, R, Casazza, V, Garavaglia, S, Ciaramella, M, Perugino, G, Rizzi, M, Rossi, F. | | 登録日 | 2013-04-02 | | 公開日 | 2013-04-17 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Biochemical and Structural Studies on the M. Tuberculosis O6-Methylguanine Methyltransferase and Mutated Variants.
J.Bacteriol., 195, 2013
|
|
8D7U
 
 | |
8D7Y
 
 | |
8D7W
 
 | |
8CVP
 
 | | Cereblon-DDB1 in the Apo form | | 分子名称: | DNA damage-binding protein 1, Protein cereblon, ZINC ION | | 著者 | Watson, E.R, Lander, G.C. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
8D7V
 
 | |
8D7X
 
 | |
8J91
 
 | | Cryo-EM structure of nucleosome containing Arabidopsis thaliana histones | | 分子名称: | DNA (169-MER), HTA13, Histone H2B.6, ... | | 著者 | Osakabe, A, Takizawa, Y, Horikoshi, N, Hatazawa, S, Berger, F, Kurumizaka, H, Kakutani, T. | | 登録日 | 2023-05-02 | | 公開日 | 2024-07-03 | | 最終更新日 | 2024-07-24 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Molecular and structural basis of the chromatin remodeling activity by Arabidopsis DDM1.
Nat Commun, 15, 2024
|
|
6NEB
 
 | | MYC Promoter G-Quadruplex with 1:6:1 loop length | | 分子名称: | DNA (27-MER) | | 著者 | Dickerhoff, J, Onel, B, Chen, L, Chen, Y, Yang, D. | | 登録日 | 2018-12-17 | | 公開日 | 2019-02-13 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Structure of a MYC Promoter G-Quadruplex with 1:6:1 Loop Length.
Acs Omega, 4, 2019
|
|
7W9V
 
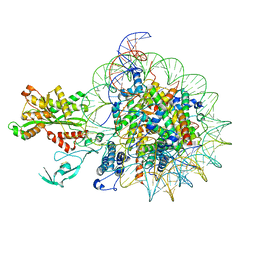 | | Cryo-EM structure of nucleosome in complex with p300 acetyltransferase catalytic core (complex I) | | 分子名称: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | 著者 | Hatazawa, S, Liu, J, Takizawa, Y, Zandian, M, Negishi, L, Kutateladze, T.G, Kurumizaka, H. | | 登録日 | 2021-12-10 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (3.95 Å) | | 主引用文献 | Structural basis for binding diversity of acetyltransferase p300 to the nucleosome.
Iscience, 25, 2022
|
|
7DBH
 
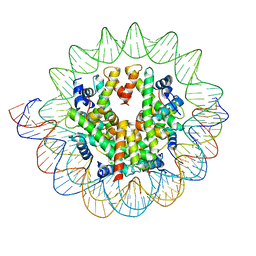 | | The mouse nucleosome structure containing H3mm18 | | 分子名称: | DNA (126-MER), Histone H2A type 1-B, Histone H2B type 3-A, ... | | 著者 | Hirai, S, Takizawa, Y, Kujirai, T, Kurumizaka, H. | | 登録日 | 2020-10-20 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Unusual nucleosome formation and transcriptome influence by the histone H3mm18 variant.
Nucleic Acids Res., 50, 2022
|
|
5Z80
 
 | | Solution structure for the 1:1 complex of a platinum(II)-based tripod bound to a hybrid-1 human telomeric G-quadruplex | | 分子名称: | 4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]-N,N-bis[4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]phenyl]aniline, G-quadruplex DNA (26-MER) | | 著者 | Liu, W.T, Zhong, Y.F, Liu, L.Y, Zeng, W.J, Wang, F.Y, Yang, D.Z, Mao, Z.W. | | 登録日 | 2018-01-30 | | 公開日 | 2018-09-19 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding
Nat Commun, 9, 2018
|
|
2AU3
 
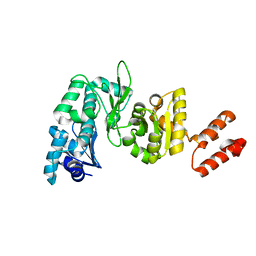 | | Crystal Structure of the Aquifex aeolicus primase (Zinc Binding and RNA Polymerase Domains) | | 分子名称: | DNA primase, ZINC ION | | 著者 | Corn, J.E, Pease, P.J, Hura, G.L, Berger, J.M. | | 登録日 | 2005-08-26 | | 公開日 | 2005-11-15 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crosstalk between primase subunits can act to regulate primer synthesis in trans.
Mol.Cell, 20, 2005
|
|
5HQ2
 
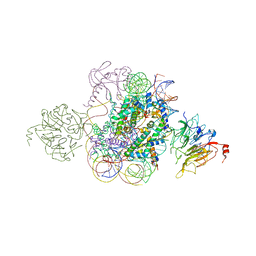 | | Structural model of Set8 histone H4 Lys20 methyltransferase bound to nucleosome core particle | | 分子名称: | DNA (149-MER), Guanine nucleotide exchange factor SRM1, Histone H2A, ... | | 著者 | Tavarekere, G, McGinty, R.K, Tan, S. | | 登録日 | 2016-01-21 | | 公開日 | 2016-03-23 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (4.5 Å) | | 主引用文献 | Multivalent Interactions by the Set8 Histone Methyltransferase With Its Nucleosome Substrate.
J.Mol.Biol., 428, 2016
|
|
8HAZ
 
 | |
1BWT
 
 | |
1BX5
 
 | |
3AIZ
 
 | |
3AIX
 
 | |
6VUG
 
 | |
1QN4
 
 | | Crystal structure of the T(-24) Adenovirus major late promoter TATA box variant bound to wild-type TBP (Arabidopsis thaliana TBP isoform 2). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. | | 分子名称: | DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*AP*AP*TP*GP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*CP*AP*TP*TP*TP*TP*AP*TP*AP*GP*C)-3'), TRANSCRIPTION INITIATION FACTOR TFIID-1 | | 著者 | Patikoglou, G.A, Kim, J.L, Sun, L, Yang, S.-H, Kodadek, T, Burley, S.K. | | 登録日 | 1999-10-14 | | 公開日 | 2000-02-06 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.86 Å) | | 主引用文献 | TATA Element Recognition by the TATA Box-Binding Protein Has Been Conserved Throughout Evolution
Genes Dev., 13, 1999
|
|
5ZB9
 
 | | Crystal structure of Rtt109 from Aspergillus fumigatus | | 分子名称: | DNA damage response protein Rtt109, putative, GLYCEROL | | 著者 | Zhang, L, Serra-Cardona, A, Zhou, H, Wang, M, Yang, N, Zhang, Z, Xu, R.M. | | 登録日 | 2018-02-10 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.502 Å) | | 主引用文献 | Multisite Substrate Recognition in Asf1-Dependent Acetylation of Histone H3 K56 by Rtt109.
Cell, 174, 2018
|
|
