8S4V
 
 | | Tankyrase 2 in complex with a quinazolin-4-one inhibitor | | 分子名称: | (2~{S})-~{N}-[2-(4-~{tert}-butylphenyl)-4-oxidanylidene-3~{H}-quinazolin-8-yl]-2,3-bis(oxidanyl)propanamide, Poly [ADP-ribose] polymerase tankyrase-2, SULFATE ION, ... | | 著者 | Bosetti, C, Lehtio, L. | | 登録日 | 2024-02-22 | | 公開日 | 2025-03-05 | | 最終更新日 | 2025-03-26 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Substitutions at the C-8 position of quinazolin-4-ones improve the potency of nicotinamide site binding tankyrase inhibitors.
Eur.J.Med.Chem., 288, 2025
|
|
8S4W
 
 | | Tankyrase 2 in complex with a quinazolin-4-one inhibitor | | 分子名称: | (2~{R})-~{N}-[2-(4-~{tert}-butylphenyl)-4-oxidanylidene-3~{H}-quinazolin-8-yl]-2,3-bis(oxidanyl)propanamide, Poly [ADP-ribose] polymerase tankyrase-2, SULFATE ION, ... | | 著者 | Bosetti, C, Lehtio, L. | | 登録日 | 2024-02-22 | | 公開日 | 2025-03-05 | | 最終更新日 | 2025-03-26 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Substitutions at the C-8 position of quinazolin-4-ones improve the potency of nicotinamide site binding tankyrase inhibitors.
Eur.J.Med.Chem., 288, 2025
|
|
6P2D
 
 | | Structure of mouse ketohexokinase-C in complex with fructose and ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Ketohexokinase, NITRATE ION, ... | | 著者 | Gasper, W.C, Allen, K.N, Tolan, D.R. | | 登録日 | 2019-05-21 | | 公開日 | 2020-06-03 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (1.79 Å) | | 主引用文献 | Michaelis-like complex of mouse ketohexokinase isoform C
ACTA CRYSTALLOGR.,SECT.D, 2024
|
|
8VG7
 
 | | Crystal Structure of T115A Variant of D-Dopachrome Tautomerase (D-DT) | | 分子名称: | D-dopachrome decarboxylase | | 著者 | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | 登録日 | 2023-12-23 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.24 Å) | | 主引用文献 | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
7Q28
 
 | |
7Q27
 
 | |
7Q29
 
 | | Crystal structure of Angiotensin-1 converting enzyme C-domain in complex with dual ACE/NEP inhibitor AD013 | | 分子名称: | (2~{S},5~{R})-5-(4-methylphenyl)-1-[2-[[(2~{S})-1-oxidanyl-1-oxidanylidene-4-phenyl-butan-2-yl]amino]ethanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Cozier, G.E, Acharya, K.R. | | 登録日 | 2021-10-23 | | 公開日 | 2022-02-16 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Probing the Requirements for Dual Angiotensin-Converting Enzyme C-Domain Selective/Neprilysin Inhibition.
J.Med.Chem., 65, 2022
|
|
8Q1V
 
 | |
7A3C
 
 | |
7OJS
 
 | |
8QMZ
 
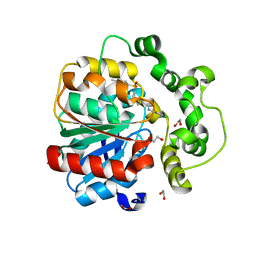 | | Soluble epoxide hydrolase in complex with RK4 | | 分子名称: | (3~{a}~{R},6~{a}~{S})-~{N}-[(2,4-dichlorophenyl)methyl]-2-(4-methylphenyl)sulfonyl-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-5-carboxamide, 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2 | | 著者 | Kumar, A, Zhu, F, Ehrler, J.M.H, Li, F, Empel, C, Xu, Y, Atodiresei, I, Koenigs, R.M, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2023-09-25 | | 公開日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Photosensitization enables Pauson-Khand-type reactions with nitrenes.
Science, 383, 2024
|
|
6UAD
 
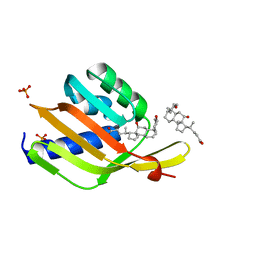 | | Ketosteroid isomerase (C. testosteroni) with truncated & designed loop for precise positioning of a catalytic E38 | | 分子名称: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, Ketosteroid isomerase with truncated and designed loop, PHOSPHATE ION | | 著者 | Kundert, K, Thompson, M.C, Liu, L, Fraser, J.S, Kortemme, T. | | 登録日 | 2019-09-10 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Ketosteroid isomerase (C. testosteroni) with truncated & designed loop for precise positioning of a catalytic E38
To Be Published
|
|
6M4P
 
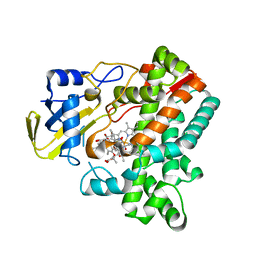 | | Cytochrome P450 monooxygenase StvP2 substrate-bound structure | | 分子名称: | 6-methoxy-streptovaricin C, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Sun, G, Hu, C, Mei, Q, Luo, M, Chen, X, Li, Z, Liu, Y, Deng, Z, Zhang, Z, Sun, Y. | | 登録日 | 2020-03-08 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Uncovering the cytochrome P450-catalyzed methylenedioxy bridge formation in streptovaricins biosynthesis.
Nat Commun, 11, 2020
|
|
3E0K
 
 | |
6PFM
 
 | | Crystal structure of GDC-0927 bound to estrogen receptor alpha | | 分子名称: | (2S)-2-(4-{2-[3-(fluoromethyl)azetidin-1-yl]ethoxy}phenyl)-3-(3-hydroxyphenyl)-4-methyl-2H-1-benzopyran-6-ol, Estrogen receptor | | 著者 | Kiefer, J.R, Vinogradova, M, Liang, J, Zhang, B, Wang, X, Zbieg, J.R, Labadie, S.S, Li, J, Ray, N.C, Ortwine, D. | | 登録日 | 2019-06-21 | | 公開日 | 2019-07-17 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.84 Å) | | 主引用文献 | Discovery of a C-8 hydroxychromene as a potent degrader of estrogen receptor alpha with improved rat oral exposure over GDC-0927.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
7SIP
 
 | | Structure of shaker-IR | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium voltage-gated channel protein Shaker | | 著者 | Tan, X, Bae, C, Stix, R, Fernandez, A.I, Huffer, K, Chang, T, Jiang, J, Faraldo-Gomez, J.D, Swartz, K.J. | | 登録日 | 2021-10-14 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of the Shaker Kv channel and mechanism of slow C-type inactivation.
Sci Adv, 8, 2022
|
|
7ACK
 
 | | CDK2/cyclin A2 in complex with an imidazo[1,2-c]pyrimidin-5-one inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 8-cyclohexyl-6~{H}-imidazo[1,2-c]pyrimidin-5-one, Cyclin-A2, ... | | 著者 | Skerlova, J, Pachl, P, Rezacova, P. | | 登録日 | 2020-09-11 | | 公開日 | 2021-03-24 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Imidazo[1,2-c]pyrimidin-5(6H)-one inhibitors of CDK2: Synthesis, kinase inhibition and co-crystal structure.
Eur.J.Med.Chem., 216, 2021
|
|
7SJ1
 
 | | Structure of shaker-W434F | | 分子名称: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, POTASSIUM ION, Potassium voltage-gated channel protein Shaker | | 著者 | Tan, X, Bae, C, Stix, R, Fernandez, A.I, Huffer, K, Chang, T, Jiang, J, Faraldo-Gomez, J.D, Swartz, K.J. | | 登録日 | 2021-10-15 | | 公開日 | 2022-03-30 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structure of the Shaker Kv channel and mechanism of slow C-type inactivation.
Sci Adv, 8, 2022
|
|
8DNQ
 
 | |
5MCS
 
 | | Solution structure and dynamics of the outer membrane cytochrome OmcF from Geobacter sulfurreducens | | 分子名称: | HEME C, Lipoprotein cytochrome c, 1 heme-binding site | | 著者 | Dantas, J.M, Silva, M.A, Morgado, L, Pantoja-Uceda, D, Turner, D.L, Bruix, M, Salgueiro, C.A. | | 登録日 | 2016-11-10 | | 公開日 | 2017-04-12 | | 最終更新日 | 2024-10-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and dynamics of the outer membrane cytochrome OmcF from Geobacter sulfurreducens.
Biochim. Biophys. Acta, 1858, 2017
|
|
4WI0
 
 | |
8CUQ
 
 | | X-ray crystal structure of ADC-33 in complex with sulfonamidoboronic acid 6e | | 分子名称: | 3-[(4R)-4-ethyl-5,7,7-trihydroxy-2,2,7-trioxo-6-oxa-2lambda~6~-thia-3-aza-7lambda~5~-phospha-5-boraheptan-1-yl]benzoic acid, Beta-lactamase | | 著者 | Fernando, M.C, Wallar, B.J, Powers, R.A. | | 登録日 | 2022-05-17 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Antibiotics, 12, 2023
|
|
8CUP
 
 | | X-ray crystal structure of ADC-33 in complex with sulfonamidoboronic acid 6d | | 分子名称: | 3-[(4S)-4-ethyl-5,7,7-trihydroxy-2,2,7-trioxo-6-oxa-2lambda~6~-thia-3-aza-7lambda~5~-phospha-5-boraheptan-1-yl]benzoic acid, Beta-lactamase | | 著者 | Fernando, M.C, Wallar, B.J, Powers, R.A. | | 登録日 | 2022-05-17 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (1.54 Å) | | 主引用文献 | Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Antibiotics, 12, 2023
|
|
8XF0
 
 | | Crystal structure of the dissociated C-phycocyanin alpha-chain from Thermoleptolyngbya sp. O-77 | | 分子名称: | C-phycocyanin alpha chain, MAGNESIUM ION, PHYCOCYANOBILIN | | 著者 | Nguyen, H.K, Teramoto, T, Kakuta, Y, Yoon, K.S. | | 登録日 | 2023-12-13 | | 公開日 | 2024-05-22 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Disassembly and reassembly of the non-conventional thermophilic C-phycocyanin.
J.Biosci.Bioeng., 137, 2024
|
|
9FEG
 
 | | PARP15 in complex with a quinazolin-4-one inhibitor | | 分子名称: | (2~{R})-~{N}-[2-(4-~{tert}-butylphenyl)-4-oxidanylidene-3~{H}-quinazolin-8-yl]-2,3-bis(oxidanyl)propanamide, (4S)-2-METHYL-2,4-PENTANEDIOL, DIMETHYL SULFOXIDE, ... | | 著者 | Bosetti, C, Lehtio, L. | | 登録日 | 2024-05-20 | | 公開日 | 2025-03-26 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Substitutions at the C-8 position of quinazolin-4-ones improve the potency of nicotinamide site binding tankyrase inhibitors.
Eur.J.Med.Chem., 288, 2025
|
|
