6RXV
 
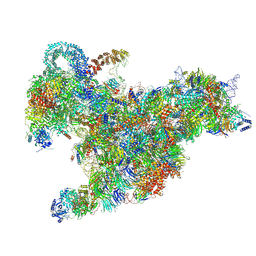 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state B2 | | 分子名称: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | 著者 | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | 登録日 | 2019-06-10 | | 公開日 | 2019-08-14 | | 最終更新日 | 2019-10-02 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6RXU
 
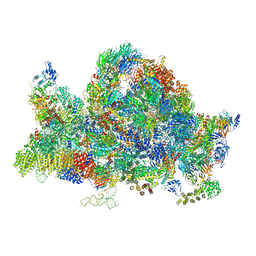 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state B1 | | 分子名称: | 35S rRNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | 著者 | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | 登録日 | 2019-06-10 | | 公開日 | 2019-08-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6OE8
 
 | | The crystal structure of hyper-thermostable AgUricase mutant K12C/E286C | | 分子名称: | MALONATE ION, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | 著者 | Shi, Y, Wang, T, Zhou, X.E, Liu, Q, Jiang, Y, Xu, H.E. | | 登録日 | 2019-03-27 | | 公開日 | 2019-08-21 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Structure-based design of a hyperthermostable AgUricase for hyperuricemia and gout therapy.
Acta Pharmacol.Sin., 40, 2019
|
|
6GEL
 
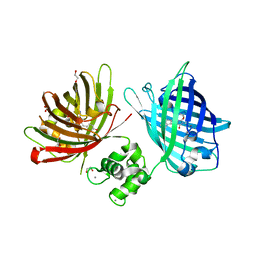 | | The structure of TWITCH-2B | | 分子名称: | CALCIUM ION, FORMIC ACID, GLYCEROL, ... | | 著者 | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | 登録日 | 2018-04-26 | | 公開日 | 2019-08-21 | | 最終更新日 | 2019-09-11 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
6GEZ
 
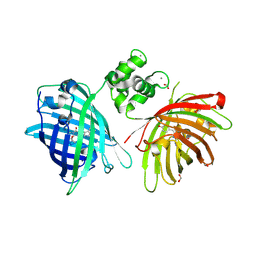 | | THE STRUCTURE OF TWITCH-2B N532F | | 分子名称: | CALCIUM ION, FORMIC ACID, Green fluorescent protein,Optimized Ratiometric Calcium Sensor,Green fluorescent protein,Green fluorescent protein | | 著者 | Trigo Mourino, P, Paulat, M, Thestrup, T, Griesbeck, O, Griesinger, C, Becker, S. | | 登録日 | 2018-04-27 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.47 Å) | | 主引用文献 | Dynamic tuning of FRET in a green fluorescent protein biosensor.
Sci Adv, 5, 2019
|
|
6HAM
 
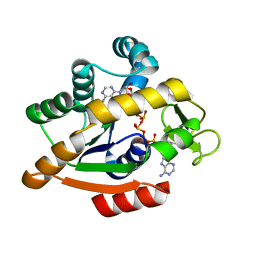 | | Adenylate kinase | | 分子名称: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE | | 著者 | Kantaev, R, Inbal, R, Goldenzweig, A, Barak, Y, Dym, O, Peleg, Y, Albek, S, Fleishman, S.J, Haran, G. | | 登録日 | 2018-08-08 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Manipulating the Folding Landscape of a Multidomain Protein.
J.Phys.Chem.B, 122, 2018
|
|
6HAP
 
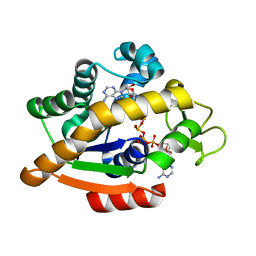 | | Adenylate kinase | | 分子名称: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE | | 著者 | Kantaev, R, Inbal, R, Goldenzweig, A, Barak, Y, Dym, O, Peleg, Y, Albek, S, Fleishman, S.J, Haran, G. | | 登録日 | 2018-08-08 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Manipulating the Folding Landscape of a Multidomain Protein.
J.Phys.Chem.B, 122, 2018
|
|
6MLT
 
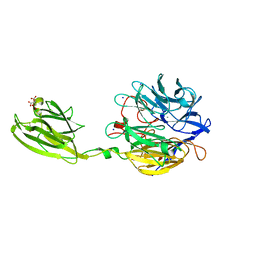 | | Crystal structure of the V. cholerae biofilm matrix protein Bap1 | | 分子名称: | CALCIUM ION, CITRATE ANION, GLYCEROL, ... | | 著者 | Kaus, K, Biester, A, Chupp, E, Lu, K, Vidsudharomn, C, Olson, R. | | 登録日 | 2018-09-28 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | The 1.9 angstrom crystal structure of the extracellular matrix protein Bap1 fromVibrio choleraeprovides insights into bacterial biofilm adhesion.
J.Biol.Chem., 294, 2019
|
|
6U95
 
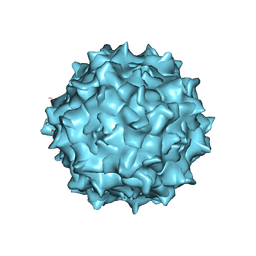 | | Adeno-associated virus strain AAVhu.37 capsid icosahedral structure | | 分子名称: | Capsid protein VP1 | | 著者 | Kaelber, J.T, Yost, S.A, Firlar, E, Mercer, A.C. | | 登録日 | 2019-09-06 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (2.56 Å) | | 主引用文献 | Structure of the AAVhu.37 capsid by cryoelectron microscopy.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6P5J
 
 | | Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 2) | | 分子名称: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | 著者 | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | 登録日 | 2019-05-30 | | 公開日 | 2019-09-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
6P4H
 
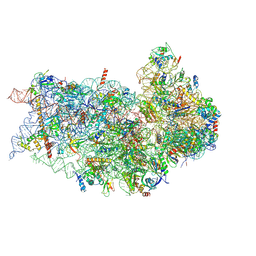 | | Structure of a mammalian small ribosomal subunit in complex with the Israeli Acute Paralysis Virus IRES (Class 2) | | 分子名称: | 18S rRNA, IAPV-IRES, RACK1, ... | | 著者 | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | 登録日 | 2019-05-27 | | 公開日 | 2019-09-18 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
6P4G
 
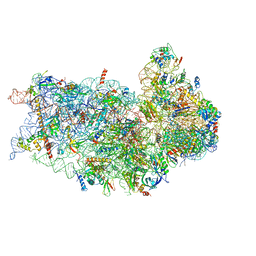 | | Structure of a mammalian small ribosomal subunit in complex with the Israeli Acute Paralysis Virus IRES (Class 1) | | 分子名称: | 18S rRNA, IAPV-IRES, RACK1, ... | | 著者 | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | 登録日 | 2019-05-27 | | 公開日 | 2019-09-18 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
6P5I
 
 | | Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 1) | | 分子名称: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | 著者 | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | 登録日 | 2019-05-30 | | 公開日 | 2019-09-18 | | 最終更新日 | 2020-02-26 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
6P5K
 
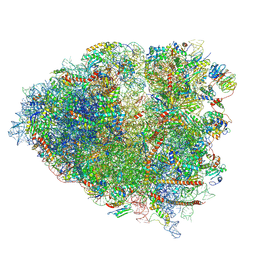 | | Structure of a mammalian 80S ribosome in complex with the Israeli Acute Paralysis Virus IRES (Class 3) | | 分子名称: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | 著者 | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | 登録日 | 2019-05-30 | | 公開日 | 2019-09-18 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
6P5N
 
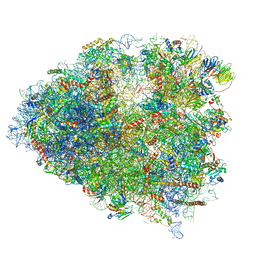 | | Structure of a mammalian 80S ribosome in complex with a single translocated Israeli Acute Paralysis Virus IRES and eRF1 | | 分子名称: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | 著者 | Acosta-Reyes, F.J, Neupane, R, Frank, J, Fernandez, I.S. | | 登録日 | 2019-05-30 | | 公開日 | 2019-09-25 | | 最終更新日 | 2020-01-08 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The Israeli acute paralysis virus IRES captures host ribosomes by mimicking a ribosomal state with hybrid tRNAs.
Embo J., 38, 2019
|
|
6RMG
 
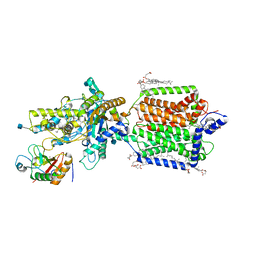 | | Structure of PTCH1 bound to a modified Hedgehog ligand ShhN-C24II | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | 著者 | Korkhov, V.M, Qi, C. | | 登録日 | 2019-05-06 | | 公開日 | 2019-10-09 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of sterol recognition by human hedgehog receptor PTCH1.
Sci Adv, 5, 2019
|
|
6MZ3
 
 | |
6MRN
 
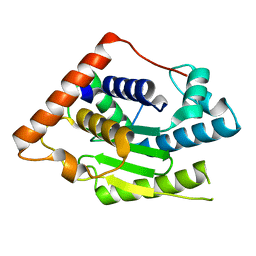 | | Crystal Structure of ChlaDUB2 DUB domain | | 分子名称: | Deubiquitinase and deneddylase Dub2 | | 著者 | Hausman, J.M, Das, C. | | 登録日 | 2018-10-15 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | The Two Deubiquitinating Enzymes fromChlamydia trachomatisHave Distinct Ubiquitin Recognition Properties.
Biochemistry, 59, 2020
|
|
6QEP
 
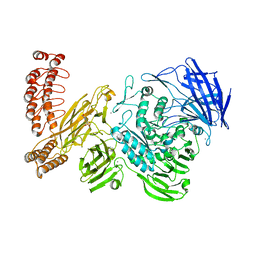 | | EngBF DARPin Fusion 4b H14 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MANGANESE (II) ION, ... | | 著者 | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | 登録日 | 2019-01-08 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
6QFO
 
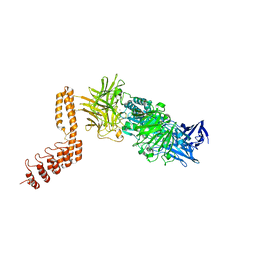 | | EngBF DARPin Fusion 9b 3G124 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, MANGANESE (II) ION, ... | | 著者 | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | 登録日 | 2019-01-10 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
6SH9
 
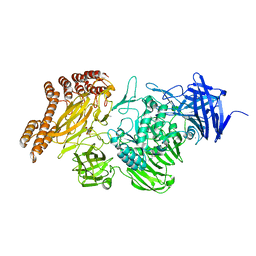 | | EngBF DARPin Fusion 4b D12 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-alpha-N-acetylgalactosaminidase,DARPin 4b D12, ... | | 著者 | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | 登録日 | 2019-08-06 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
6QEV
 
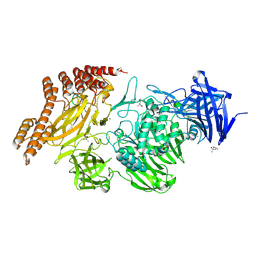 | | EngBF DARPin Fusion 4b B6 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, MANGANESE (II) ION, PEGA domain-containing protein,PEGA domain-containing protein,EngBF DARPin fusion B6 complex, ... | | 著者 | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | 登録日 | 2019-01-08 | | 公開日 | 2019-11-06 | | 最終更新日 | 2020-04-22 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
6QFK
 
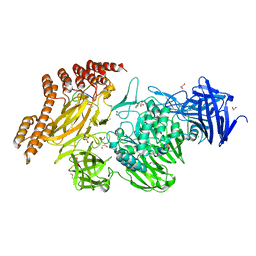 | | EngBF DARPin Fusion 4b G10 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | 著者 | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | 登録日 | 2019-01-10 | | 公開日 | 2019-11-06 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
6IR2
 
 | | Crystal structure of red fluorescent protein mCherry complexed with the nanobody LaM2 at 1.4 Angstron resolution | | 分子名称: | MCherry fluorescent protein, mCherry's nanobody LaM2 | | 著者 | Ding, Y, Wang, Z.Y, Hu, R.T, Chen, X. | | 登録日 | 2018-11-09 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.393 Å) | | 主引用文献 | Structural insights into the binding of nanobodies LaM2 and LaM4 to the red fluorescent protein mCherry.
Protein Sci., 30, 2021
|
|
6GOZ
 
 | | Structure of mEos4b in the green long-lived dark state | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | De Zitter, E, Adam, V, Byrdin, M, Van Meervelt, L, Dedecker, P, Bourgeois, D. | | 登録日 | 2018-06-04 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.406 Å) | | 主引用文献 | Mechanistic Investigations of Green mEos4b Reveal a Dynamic Long-Lived Dark State.
J.Am.Chem.Soc., 2020
|
|
