5JOX
 
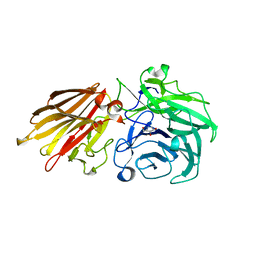 | | Bacteroides ovatus Xyloglucan PUL GH43A in complex with AraDNJ | | 分子名称: | 1,4-DIDEOXY-1,4-IMINO-L-ARABINITOL, Non-reducing end alpha-L-arabinofuranosidase BoGH43A | | 著者 | Thompson, A.J, Hemsworth, G.R, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | 登録日 | 2016-05-03 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
1RWP
 
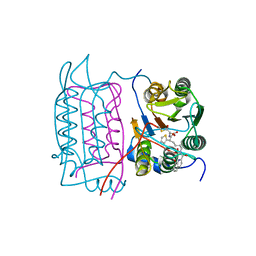 | |
1BSC
 
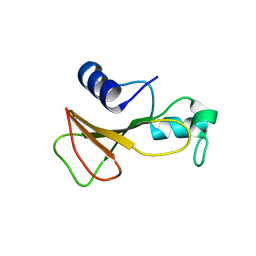 | |
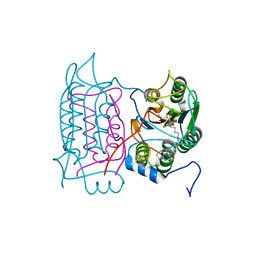
1RWX
 
 | |
1BSX
 
 | | STRUCTURE AND SPECIFICITY OF NUCLEAR RECEPTOR-COACTIVATOR INTERACTIONS | | 分子名称: | 3,5,3'TRIIODOTHYRONINE, PROTEIN (GRIP1), PROTEIN (THYROID HORMONE RECEPTOR BETA) | | 著者 | Wagner, R.L, Darimont, B.D, Apriletti, J.W, Stallcup, M.R, Kushner, P.J, Baxter, J.D, Fletterick, R.J, Yamamoto, K.R. | | 登録日 | 1998-08-31 | | 公開日 | 1999-08-26 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | Structure and specificity of nuclear receptor-coactivator interactions.
Genes Dev., 12, 1998
|
|
3K7G
 
 | | Crystal structure of the Indian Hedgehog N-terminal signalling domain | | 分子名称: | GLYCEROL, Indian hedgehog protein, SULFATE ION, ... | | 著者 | He, Y.-X, Kang, Y, Zhang, W.J, Yu, J, Ma, G, Zhou, C.-Z. | | 登録日 | 2009-10-13 | | 公開日 | 2011-01-26 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structure of the Indian Hedgehog N-terminal signalling domain
To be Published
|
|
1S7Q
 
 | | Crystal structures of the murine class I major histocompatibility complex H-2Kb in complex with LCMV-derived gp33 index peptide and three of its escape variants | | 分子名称: | Beta-2-microglobulin, Glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | 著者 | Velloso, L.M, Michaelsson, J, Ljunggren, H.G, Schneider, G, Achour, A. | | 登録日 | 2004-01-30 | | 公開日 | 2004-05-04 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Determination of structural principles underlying three different modes of lymphocytic choriomeningitis virus escape from CTL recognition.
J.Immunol., 172, 2004
|
|
1RZT
 
 | | Crystal structure of DNA polymerase lambda complexed with a two nucleotide gap DNA molecule | | 分子名称: | 1,2-ETHANEDIOL, 5'-D(*CP*GP*GP*CP*AP*AP*CP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*G)-3', ... | | 著者 | Pedersen, L.C, Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Blanco, L, Kunkel, T.A. | | 登録日 | 2003-12-29 | | 公開日 | 2004-03-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology.
Mol.Cell, 13, 2004
|
|
1S7X
 
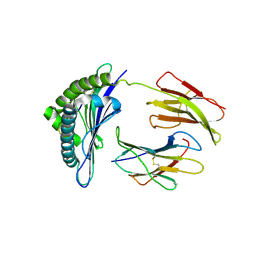 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 index peptide and three of its escape variants | | 分子名称: | Beta-2-microglobulin, Glycoprotein 9-residue peptide, H-2 class I histocompatibility antigen, ... | | 著者 | Velloso, L.M, Michaelsson, J, Ljunggren, H.G, Schneider, G, Achour, A. | | 登録日 | 2004-01-30 | | 公開日 | 2004-05-04 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Determination of structural principles underlying three different modes of lymphocytic choriomeningitis virus escape from CTL recognition.
J.Immunol., 172, 2004
|
|
5SVN
 
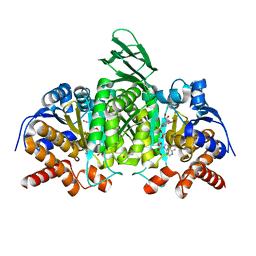 | | Structure of IDH2 mutant R172K | | 分子名称: | DI(HYDROXYETHYL)ETHER, Isocitrate dehydrogenase [NADP], mitochondrial, ... | | 著者 | Xie, X, Kulathila, R. | | 登録日 | 2016-08-06 | | 公開日 | 2017-02-08 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Allosteric Mutant IDH1 Inhibitors Reveal Mechanisms for IDH1 Mutant and Isoform Selectivity.
Structure, 25, 2017
|
|
5SVZ
 
 | |
1CB0
 
 | |
5JRT
 
 | |
5SV7
 
 | | The Crystal structure of a chaperone | | 分子名称: | Eukaryotic translation initiation factor 2-alpha kinase 3 | | 著者 | Wang, P, Li, J, Sha, B. | | 登録日 | 2016-08-04 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.209 Å) | | 主引用文献 | The ER stress sensor PERK luminal domain functions as a molecular chaperone to interact with misfolded proteins.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1SC4
 
 | |
5TC8
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with methylthio-DADMe-Immucillin-A | | 分子名称: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | 登録日 | 2016-09-14 | | 公開日 | 2017-10-11 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | TBA
To be published
|
|
5JXD
 
 | | Crystal structure of murine Tnfaip8 C165S mutant | | 分子名称: | Tumor necrosis factor alpha-induced protein 8, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | 著者 | Park, J, Kim, M.S, Lee, D, Shin, D.H. | | 登録日 | 2016-05-13 | | 公開日 | 2017-06-28 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.029 Å) | | 主引用文献 | The Tnfaip8-PE complex is a novel upstream effector in the anti-autophagic action of insulin
Sci Rep, 7, 2017
|
|
3KA0
 
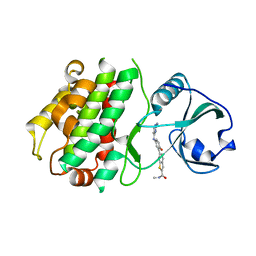 | |
4X5V
 
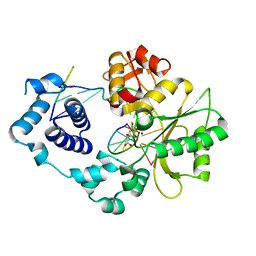 | | Crystal structure of the post-catalytic nick complex of DNA polymerase lambda with a templating A and incorporated 8-oxo-dGMP | | 分子名称: | CITRIC ACID, DNA (5'-D(*CP*AP*GP*TP*AP*CP*(8OG))-3'), DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*AP*CP*TP*G)-3'), ... | | 著者 | Burak, M.J, Guja, K.E, Garcia-Diaz, M. | | 登録日 | 2014-12-05 | | 公開日 | 2015-10-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Nucleotide binding interactions modulate dNTP selectivity and facilitate 8-oxo-dGTP incorporation by DNA polymerase lambda.
Nucleic Acids Res., 43, 2015
|
|
4X61
 
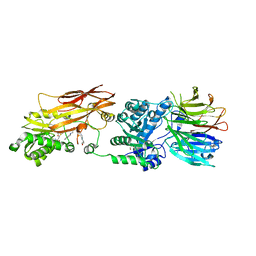 | | Crystal structure of PRMT5:MEP50 with EPZ015666 and SAM | | 分子名称: | GLYCEROL, Methylosome protein 50, N-[(2S)-3-(3,4-dihydroisoquinolin-2(1H)-yl)-2-hydroxypropyl]-6-(oxetan-3-ylamino)pyrimidine-4-carboxamide, ... | | 著者 | Boriack-Sjodin, P.A. | | 登録日 | 2014-12-06 | | 公開日 | 2015-04-22 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models.
Nat.Chem.Biol., 11, 2015
|
|
1U0F
 
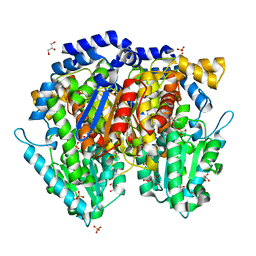 | | Crystal structure of mouse phosphoglucose isomerase in complex with glucose 6-phosphate | | 分子名称: | 6-O-phosphono-alpha-D-glucopyranose, BETA-MERCAPTOETHANOL, GLUCOSE-6-PHOSPHATE, ... | | 著者 | Solomons, J.T.G, Zimmerly, E.M, Burns, S, Krishnamurthy, N, Swan, M.K, Krings, S, Muirhead, H, Chirgwin, J, Davies, C. | | 登録日 | 2004-07-13 | | 公開日 | 2004-11-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | The crystal structure of mouse phosphoglucose isomerase at 1.6A resolution and its complex with glucose 6-phosphate reveals the catalytic mechanism of sugar ring opening.
J.Mol.Biol., 342, 2004
|
|
3JS5
 
 | |
3K23
 
 | | Glucocorticoid Receptor with Bound D-prolinamide 11 | | 分子名称: | 1-{[3-(4-{[(2R)-4-(5-fluoro-2-methoxyphenyl)-2-hydroxy-4-methyl-2-(trifluoromethyl)pentyl]amino}-6-methyl-1H-indazol-1-yl)phenyl]carbonyl}-D-prolinamide, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | 著者 | Biggadike, K.B, McLay, I.M, Madauss, K.P, Williams, S.P, Bledsoe, R.K. | | 登録日 | 2009-09-29 | | 公開日 | 2009-10-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Design and x-ray crystal structures of high-potency nonsteroidal glucocorticoid agonists exploiting a novel binding site on the receptor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5TM3
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with 2,3-bis(2-chloro-4-hydroxyphenyl)thiophene 1-oxide | | 分子名称: | (1S)-2,3-bis(2-chloro-4-hydroxyphenyl)-1H-1lambda~4~-thiophen-1-one, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-12 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.194 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TML
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, (E)-6-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenyl)hex-5-enoic acid | | 分子名称: | 6-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenyl}hex-5-enoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | 著者 | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | 登録日 | 2016-10-13 | | 公開日 | 2017-01-18 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
