1S9G
 
 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R120394. | | 分子名称: | 4-[4-AMINO-6-(5-CHLORO-1H-INDOL-4-YLMETHYL)-[1,3,5]TRIAZIN-2-YLAMINO]-BENZONITRILE, POL polyprotein [Contains: Reverse transcriptase] | | 著者 | Das, K, Clark Jr, A.D, Ludovici, D.W, Kukla, M.J, Decorte, B, Lewi, P.J, Hughes, S.H, Janssen, P.A, Arnold, E. | | 登録日 | 2004-02-04 | | 公開日 | 2004-05-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants.
J.Med.Chem., 47, 2004
|
|
5T1V
 
 | | Crystal structure of Zika virus NS2B-NS3 protease in apo-form. | | 分子名称: | NS2B-NS3 protease,NS2B-NS3 protease | | 著者 | Nocadello, S, Light, S.H, Minasov, G, Shuvalova, L, Cardona-Correa, A.A, Ojeda, I, Vargas, J, Johnson, M.E, Lee, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2016-08-22 | | 公開日 | 2016-09-07 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structure of Zika virus NS2B-NS3 protease in apo-form.
To Be Published
|
|
8GUB
 
 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant H1047R in complex with BYL-719 | | 分子名称: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | 著者 | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | 登録日 | 2022-09-11 | | 公開日 | 2022-11-23 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (2.73 Å) | | 主引用文献 | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6GYU
 
 | | Cryo-EM structure of the CBF3-msk complex of the budding yeast kinetochore | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-02 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4AAC
 
 | | P38ALPHA MAP KINASE BOUND TO CMPD 29 | | 分子名称: | CHLORIDE ION, MITOGEN-ACTIVATED PROTEIN KINASE 14, N-isoxazol-3-yl-4-methyl-3-[6-(4-methylpiperazin-1-yl)-4-oxo-quinazolin-3-yl]benzamide | | 著者 | Gerhardt, S, Hargreaves, D. | | 登録日 | 2011-12-01 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The Discovery of N-Cyclopropyl-4-Methyl-3-[6--(4-Methylpiperazin-1-Yl-4-Oxoquinazolin-3(4H)-Yl]Benzamide (Azd6703), a Clinical P38Alpha Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6G3Y
 
 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH5675 | | 分子名称: | 4-(4-azanyl-2-oxidanylidene-3~{H}-benzimidazol-1-yl)-~{N}-(4-iodophenyl)piperidine-1-carboxamide, ACETATE ION, N-glycosylase/DNA lyase, ... | | 著者 | Masuyer, G, Helleday, T, Stenmark, P. | | 登録日 | 2018-03-26 | | 公開日 | 2018-11-28 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.51 Å) | | 主引用文献 | Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation.
Science, 362, 2018
|
|
1RT3
 
 | | AZT DRUG RESISTANT HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH 1051U91 | | 分子名称: | 6,11-DIHYDRO-11-ETHYL-6-METHYL-9-NITRO-5H-PYRIDO[2,3-B][1,5]BENZODIAZEPIN-5-ONE, HIV-1 REVERSE TRANSCRIPTASE | | 著者 | Ren, J, Stammers, D.K, Stuart, D.I. | | 登録日 | 1998-06-29 | | 公開日 | 1999-02-16 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | 3'-Azido-3'-deoxythymidine drug resistance mutations in HIV-1 reverse transcriptase can induce long range conformational changes.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1RTJ
 
 | | MECHANISM OF INHIBITION OF HIV-1 REVERSE TRANSCRIPTASE BY NON-NUCLEOSIDE INHIBITORS | | 分子名称: | HIV-1 REVERSE TRANSCRIPTASE | | 著者 | Ren, J, Esnouf, R, Ross, C, Jones, Y, Stammers, D, Stuart, D. | | 登録日 | 1995-05-03 | | 公開日 | 1996-04-03 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Mechanism of inhibition of HIV-1 reverse transcriptase by non-nucleoside inhibitors.
Nat.Struct.Biol., 2, 1995
|
|
6GYS
 
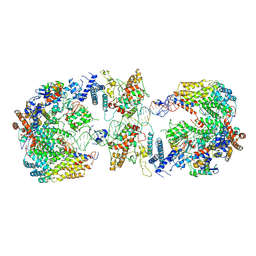 | | Cryo-EM structure of the CBF3-CEN3 complex of the budding yeast kinetochore | | 分子名称: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-01 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-11 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6GYP
 
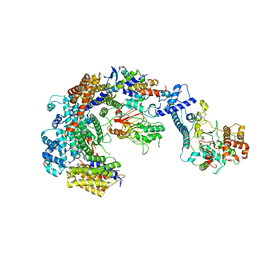 | | Cryo-EM structure of the CBF3-core-Ndc10-DBD complex of the budding yeast kinetochore | | 分子名称: | ARGININE, ASPARAGINE, Centromere DNA-binding protein complex CBF3 subunit A, ... | | 著者 | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | 登録日 | 2018-07-01 | | 公開日 | 2018-12-05 | | 最終更新日 | 2019-12-18 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3UVN
 
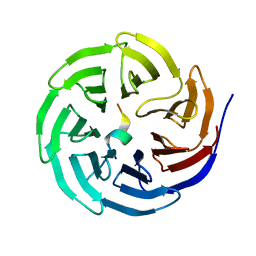 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of SET1A | | 分子名称: | Histone-lysine N-methyltransferase SETD1A, WD repeat-containing protein 5 | | 著者 | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | 登録日 | 2011-11-30 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.792 Å) | | 主引用文献 | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
3UVM
 
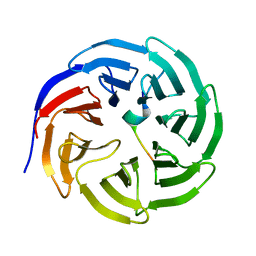 | | Crystal structure of WDR5 in complex with the WDR5-interacting motif of MLL4 | | 分子名称: | Histone-lysine N-methyltransferase MLL4, WD repeat-containing protein 5 | | 著者 | Zhang, P, Lee, H, Brunzelle, J.S, Couture, J.-F. | | 登録日 | 2011-11-30 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | The plasticity of WDR5 peptide-binding cleft enables the binding of the SET1 family of histone methyltransferases.
Nucleic Acids Res., 40, 2012
|
|
1EBM
 
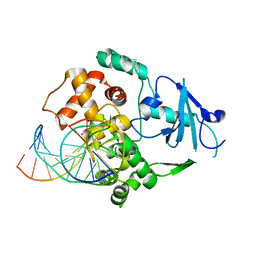 | | CRYSTAL STRUCTURE OF THE HUMAN 8-OXOGUANINE GLYCOSYLASE (HOGG1) BOUND TO A SUBSTRATE OLIGONUCLEOTIDE | | 分子名称: | 8-OXOGUANINE DNA GLYCOSYLASE, CALCIUM ION, DNA (5'-D(*GP*CP*GP*TP*CP*CP*AP*(8OG)P*GP*TP*CP*TP*AP*CP*C)-3'), ... | | 著者 | Bruner, S.D, Norman, D.P, Verdine, G.L. | | 登録日 | 2000-01-24 | | 公開日 | 2000-03-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA.
Nature, 403, 2000
|
|
1SUQ
 
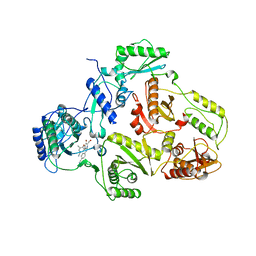 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R185545 | | 分子名称: | (6-[4-(AMINOMETHYL)-2,6-DIMETHYLPHENOXY]-2-{[4-(AMINOMETHYL)PHENYL]AMINO}-5-BROMOPYRIMIDIN-4-YL)METHANOL, MAGNESIUM ION, REVERSE TRANSCRIPTASE | | 著者 | Das, K, Arnold, E. | | 登録日 | 2004-03-26 | | 公開日 | 2004-05-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
3UR4
 
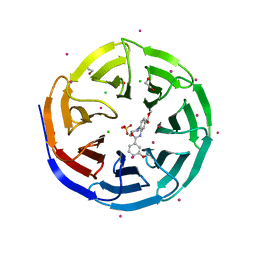 | | Crystal structure of human WD repeat domain 5 with compound | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, SULFATE ION, ... | | 著者 | Dong, A, Dombrovski, L, Senisterra, G, Wernimont, A, Wasney, G.A, Allali Hassani, A, Nguyen, K.T, Smil, D, Bolshan, Y, Hajian, T, Poda, G, Chau, I, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Brown, P, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | 登録日 | 2011-11-21 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
3UV4
 
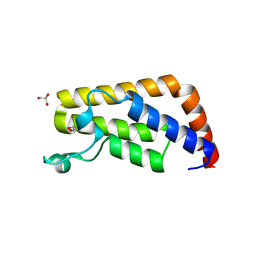 | | Crystal Structure of the second bromodomain of human Transcription initiation factor TFIID subunit 1 (TAF1) | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHATE ION, ... | | 著者 | Filippakopoulos, P, Picaud, S, Keates, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | 登録日 | 2011-11-29 | | 公開日 | 2012-03-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
7CFQ
 
 | | Crystal structure of WDR5 in complex with H3K4me3Q5ser peptide | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, H3K4me3Q5ser peptide, ... | | 著者 | Zhao, J, Zhang, X, Zang, J. | | 登録日 | 2020-06-27 | | 公開日 | 2021-07-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural insights into the recognition of histone H3Q5 serotonylation by WDR5.
Sci Adv, 7, 2021
|
|
7CJB
 
 | |
4KIL
 
 | | 7-(4-fluorophenyl)-3-hydroxyquinolin-2(1H)-one bound to influenza 2009 H1N1 endonuclease | | 分子名称: | 1,2-ETHANEDIOL, 7-(4-fluorophenyl)-3-hydroxyquinolin-2(1H)-one, MANGANESE (II) ION, ... | | 著者 | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | 登録日 | 2013-05-02 | | 公開日 | 2013-05-29 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | 3-Hydroxyquinolin-2(1H)-ones As Inhibitors of Influenza A Endonuclease.
ACS Med Chem Lett, 4, 2013
|
|
1SV5
 
 | | CRYSTAL STRUCTURE OF K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R165335 | | 分子名称: | 4-({6-AMINO-5-BROMO-2-[(4-CYANOPHENYL)AMINO]PYRIMIDIN-4-YL}OXY)-3,5-DIMETHYLBENZONITRILE, Reverse Transcriptase | | 著者 | Das, K, Arnold, E. | | 登録日 | 2004-03-27 | | 公開日 | 2004-05-11 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
4EH3
 
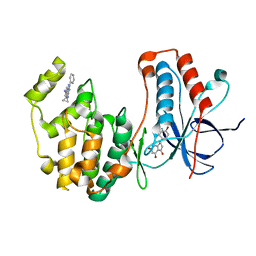 | | Human p38 MAP kinase in complex with NP-F2 and RL87 | | 分子名称: | Mitogen-activated protein kinase 14, NARINGENIN, N~4~-cyclopropyl-2-phenylquinazoline-4,7-diamine | | 著者 | Over, B, Gruetter, C, Waldmann, H, Rauh, D. | | 登録日 | 2012-04-02 | | 公開日 | 2012-12-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Natural-product-derived fragments for fragment-based ligand discovery.
Nat Chem, 5, 2012
|
|
4EH6
 
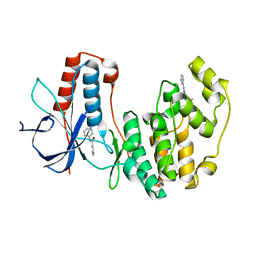 | | Human p38 MAP kinase in complex with NP-F5 and RL87 | | 分子名称: | Mitogen-activated protein kinase 14, N-phenylpyridine-3-carboxamide, N~4~-cyclopropyl-2-phenylquinazoline-4,7-diamine | | 著者 | Over, B, Gruetter, C, Waldmann, H, Rauh, D. | | 登録日 | 2012-04-02 | | 公開日 | 2012-12-05 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Natural-product-derived fragments for fragment-based ligand discovery.
Nat Chem, 5, 2012
|
|
1TQE
 
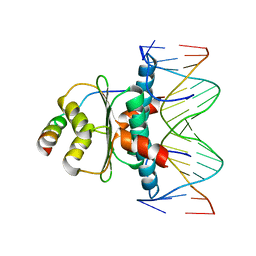 | | Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2 | | 分子名称: | Histone deacetylase 9, MEF2 binding site of nur77 promoter, Myocyte-specific enhancer factor 2B | | 著者 | Chen, L, Han, A, He, J, Wu, Y, Liu, J.O. | | 登録日 | 2004-06-17 | | 公開日 | 2004-12-21 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Mechanism of Recruitment of Class II Histone Deacetylases by Myocyte Enhancer Factor-2.
J.Mol.Biol., 345, 2005
|
|
6U62
 
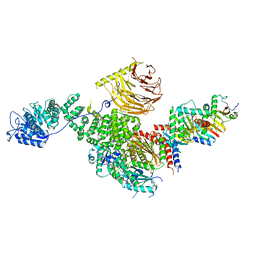 | | Raptor-Rag-Ragulator complex | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Rogala, K.B, Sabatini, D.M. | | 登録日 | 2019-08-29 | | 公開日 | 2019-10-30 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.18 Å) | | 主引用文献 | Structural basis for the docking of mTORC1 on the lysosomal surface.
Science, 366, 2019
|
|
4IZ7
 
 | |
