7U3T
 
 | | [F223] Self-assembling tensegrity triangle with two turns, two turns and three turns of DNA per axis by extension with P1 symmetry | | 分子名称: | DNA (31-MER), DNA (5'-D(*AP*AP*CP*CP*TP*AP*CP*CP*TP*GP*GP*CP*AP*GP*GP*AP*CP*GP*AP*CP*T)-3'), DNA (5'-D(*CP*AP*CP*GP*AP*GP*CP*CP*TP*GP*AP*TP*CP*GP*GP*AP*CP*AP*AP*GP*A)-3'), ... | | 著者 | Woloszyn, K, Vecchioni, S, Seeman, N.C, Sha, R, Ohayon, Y.P, Zhu, E. | | 登録日 | 2022-02-28 | | 公開日 | 2022-09-28 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (4.69 Å) | | 主引用文献 | Augmented DNA Nanoarchitectures: A Structural Library of 3D Self-Assembling Tensegrity Triangle Variants.
Adv Mater, 34, 2022
|
|
8GUV
 
 | | LecA from Pseudomonas aeruginosa in complex with tolcapone (CAS: 134308-13-7) | | 分子名称: | CALCIUM ION, PA-I galactophilic lectin, Tolcapone | | 著者 | Kuhaudomlarp, S, Siebs, E, Varrot, A, Imberty, A, Titz, A. | | 登録日 | 2022-09-13 | | 公開日 | 2023-07-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | LecA from Pseudomonas aeruginosa in complex with tolcapone (CAS: 134308-13-7)
To Be Published
|
|
8G7J
 
 | | mtHsp60 V72I apo | | 分子名称: | 60 kDa heat shock protein, mitochondrial | | 著者 | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | 登録日 | 2023-02-16 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7K
 
 | | mtHsp60 V72I apo focus | | 分子名称: | 60 kDa heat shock protein, mitochondrial | | 著者 | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | 登録日 | 2023-02-16 | | 公開日 | 2023-07-12 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8GAK
 
 | | Crystal Structure of E. coli LptA in complex with Chinavia Ubica Thanatin | | 分子名称: | Lipopolysaccharide export system protein LptA, Thanatin | | 著者 | Huynh, K, Kibrom, A, Donald, B, Zhou, P. | | 登録日 | 2023-02-23 | | 公開日 | 2023-07-19 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Discovery, characterization, and redesign of potent antimicrobial thanatin orthologs from Chinavia ubica and Murgantia histrionica targeting E. coli LptA.
J Struct Biol X, 8, 2023
|
|
7SQR
 
 | | 201phi2-1 Chimallin localized tetramer reconstruction | | 分子名称: | Chimallin | | 著者 | Laughlin, T.G, Deep, A, Prichard, A.M, Seitz, C, Gu, Y, Enustun, E, Suslov, S, Khanna, K, Birkholz, E.A, Amaro, R.E, Pogliano, J, Corbett, K.D, Villa, E. | | 登録日 | 2021-11-06 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Architecture and self-assembly of the jumbo bacteriophage nuclear shell.
Nature, 608, 2022
|
|
7A1U
 
 | | Structure of SARS-CoV-2 Main Protease bound to Fusidic Acid. | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, FUSIDIC ACID, ... | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-08-14 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7SQU
 
 | | Goslar chimallin C4 tetramer localized reconstruction | | 分子名称: | Chimallin | | 著者 | Laughlin, T.G, Deep, A, Prichard, A.M, Seitz, C, Gu, Y, Enustun, E, Suslov, S, Khanna, K, Birkholz, E.A, Amaro, R.E, Pogliano, J, Corbett, K.D, Villa, E. | | 登録日 | 2021-11-06 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (2.6 Å) | | 主引用文献 | Architecture and self-assembly of the jumbo bacteriophage nuclear shell.
Nature, 608, 2022
|
|
7U4R
 
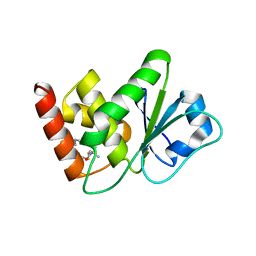 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((9-propyl-5,6-dihydrothieno[2,3-h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | 分子名称: | 3,3-dimethyl-1-{[(9aM)-9-propyl-5,6-dihydrothieno[2,3-h]quinazolin-2-yl]sulfanyl}butan-2-one, Dual specificity protein phosphatase 10 | | 著者 | Gannam, Z.T.K, Jamali, H, Gentzel, E, Lolis, E, Anderson, K.S, Ellman, J.A, Bennett, A.M. | | 登録日 | 2022-02-28 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.14 Å) | | 主引用文献 | Defining the structure-activity relationship for a novel class of allosteric MKP5 inhibitors.
Eur.J.Med.Chem., 243, 2022
|
|
7A09
 
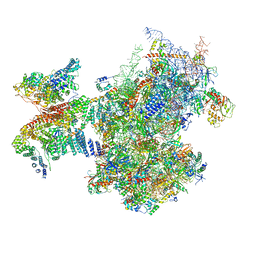 | | Structure of a human ABCE1-bound 43S pre-initiation complex - State III | | 分子名称: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | 著者 | Kratzat, H, Mackens-Kiani, T, Ameismeier, A, Cheng, J, Berninghausen, O, Becker, T, Beckmann, R. | | 登録日 | 2020-08-07 | | 公開日 | 2020-10-14 | | 最終更新日 | 2021-01-13 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | A structural inventory of native ribosomal ABCE1-43S pre-initiation complexes.
Embo J., 40, 2021
|
|
7A2D
 
 | | Structure-function analyses of dual-BON domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation to the cell division site | | 分子名称: | Uncharacterized protein YraP | | 著者 | Bryant, J.A, Morris, F.C, Knowles, T.J, Maderbocus, R, Heinz, E, Boelter, G, Alodaini, D, Colyer, A, Wotherspoon, P.J, Staunton, K.A, Jeeves, M, Browning, D.F, Sevastsyanovich, Y.R, Wells, T.J, Rossiter, A.E, Bavro, V.N, Sridhar, P, Ward, D.G, Chong, Z.S, Goodall, E.C.A, Icke, C, Teo, A, Chng, S.S, Roper, D.I, Lithgow, T, Cunningham, A.F, Banzhaf, M, Overduin, M, Henderson, I.R. | | 登録日 | 2020-08-17 | | 公開日 | 2020-12-30 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of dual BON-domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation.
Elife, 9, 2020
|
|
5N9R
 
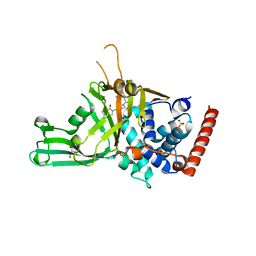 | | Crystal structure of USP7 in complex with a potent, selective and reversible small-molecule inhibitor | | 分子名称: | 7-bromanyl-3-[[4-oxidanyl-1-[(3~{R})-3-phenylbutanoyl]piperidin-4-yl]methyl]thieno[3,2-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | 著者 | Harrison, T, Gavory, G, O'Dowd, C, Helm, M, Flasz, I, Arkoudis, E, Dossang, A, Hughes, C, Cassidy, E, McClelland, K, Odrzywol, E, Page, N, Barker, O, Miel, H. | | 登録日 | 2017-02-27 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Discovery and characterization of highly potent and selective allosteric USP7 inhibitors.
Nat. Chem. Biol., 14, 2018
|
|
7RFU
 
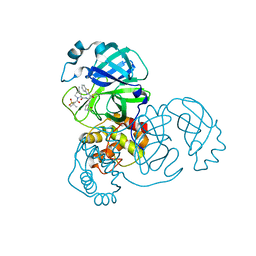 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | 分子名称: | (1R,2S,5S)-N-{(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-3-[N-(methanesulfonyl)-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | 登録日 | 2021-07-14 | | 公開日 | 2021-11-10 | | 最終更新日 | 2022-01-05 | | 実験手法 | X-RAY DIFFRACTION (2.498 Å) | | 主引用文献 | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
7RFW
 
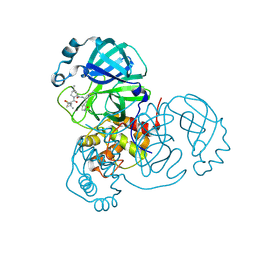 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | 登録日 | 2021-07-14 | | 公開日 | 2021-11-10 | | 最終更新日 | 2022-01-05 | | 実験手法 | X-RAY DIFFRACTION (1.729 Å) | | 主引用文献 | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
7RFS
 
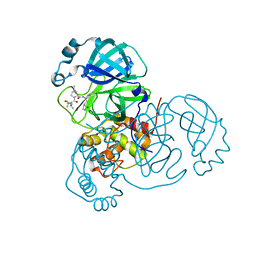 | | Structure of SARS-CoV-2 main protease in complex with a covalent inhibitor | | 分子名称: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | 著者 | Greasley, S.E, Ferre, R.A, Liu, W, Stewart, A.E. | | 登録日 | 2021-07-14 | | 公開日 | 2021-11-10 | | 最終更新日 | 2022-01-05 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | An oral SARS-CoV-2 M pro inhibitor clinical candidate for the treatment of COVID-19.
Science, 374, 2021
|
|
6ZQ4
 
 | |
6FQM
 
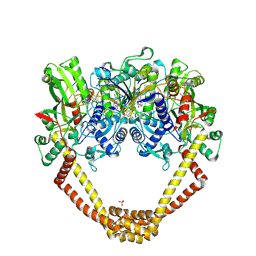 | | 3.06A COMPLEX OF S.AUREUS GYRASE with imidazopyrazinone T1 AND DNA | | 分子名称: | 7-[(3~{S})-3-azanylpyrrolidin-1-yl]-5-cyclopropyl-8-fluoranyl-imidazo[1,2-a]quinoxalin-4-one, DNA (5'-D(*GP*AP*GP*AP*GP*TP*AP*T*GP*GP*CP*CP*AP*TP*AP*CP*TP*CP*T)-3'), DNA gyrase subunit A, ... | | 著者 | Bax, B.D, Germe, T, Basque, E, Maxwell, A. | | 登録日 | 2018-02-14 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.06 Å) | | 主引用文献 | A new class of antibacterials, the imidazopyrazinones, reveal structural transitions involved in DNA gyrase poisoning and mechanisms of resistance.
Nucleic Acids Res., 46, 2018
|
|
6VS7
 
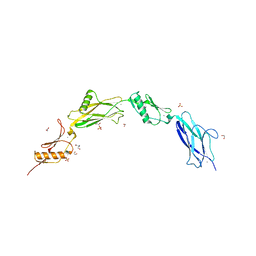 | | Sialic acid binding region of Streptococcus Sanguinis SK1 adhesin | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Adhesin, ... | | 著者 | Stubbs, H.E, Iverson, T.M. | | 登録日 | 2020-02-10 | | 公開日 | 2020-08-26 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Tandem sialoglycan-binding modules in a Streptococcus sanguinis serine-rich repeat adhesin create target dependent avidity effects.
J.Biol.Chem., 295, 2020
|
|
6BHJ
 
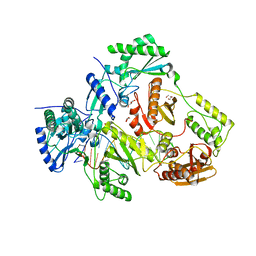 | | Structure of HIV-1 Reverse Transcriptase Bound to a 38-mer Hairpin Template-Primer RNA-DNA Aptamer | | 分子名称: | 38-MER RNA-DNA Aptamer, GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | 著者 | Ruiz, F.X, Miller, M.T, Tuske, S, Das, K, Arnold, E. | | 登録日 | 2017-10-30 | | 公開日 | 2018-10-31 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | Integrative Structural Biology Studies of HIV-1 Reverse Transcriptase Binding to a High-Affinity DNA Aptamer
Curr Res Struct Biol, 2020
|
|
6FQV
 
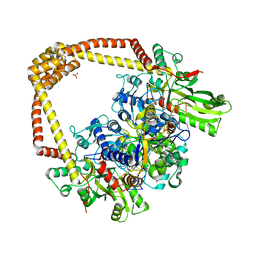 | | 2.60A BINARY COMPLEX OF S.AUREUS GYRASE with UNCLEAVED DNA | | 分子名称: | DNA (5'-D(*GP*AP*GP*CP*GP*TP*AP*CP*GP*GP*CP*CP*GP*TP*AP*CP*GP*CP*TP*T)-3'), DNA gyrase subunit A, DNA gyrase subunit B,DNA gyrase subunit B, ... | | 著者 | Bax, B.D, Germe, T, Basque, E, Maxwell, A. | | 登録日 | 2018-02-14 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A new class of antibacterials, the imidazopyrazinones, reveal structural transitions involved in DNA gyrase poisoning and mechanisms of resistance.
Nucleic Acids Res., 46, 2018
|
|
8UU7
 
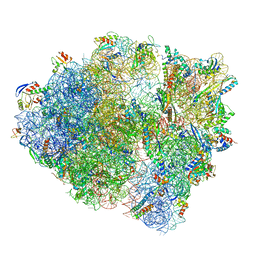 | | Cryo-EM structure of the Listeria innocua 70S ribosome in complex with HflXr, HPF, and E-site tRNA (structure II-B) | | 分子名称: | 16S Ribosomal RNA, 23S Ribosomal RNA, 5S Ribosomal RNA, ... | | 著者 | Seely, S.M, Basu, R.S, Gagnon, M.G. | | 登録日 | 2023-10-31 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Mechanistic insights into the alternative ribosome recycling by HflXr.
Nucleic Acids Res., 52, 2024
|
|
5N9T
 
 | | Crystal structure of USP7 in complex with a potent, selective and reversible small-molecule inhibitor | | 分子名称: | 3-[4-(aminomethyl)phenyl]-2-methyl-6-[[4-oxidanyl-1-[(3~{R})-4,4,4-tris(fluoranyl)-3-phenyl-butanoyl]piperidin-4-yl]methyl]pyrazolo[4,3-d]pyrimidin-7-one, GLYCEROL, SULFATE ION, ... | | 著者 | Harrison, T, Gavory, G, O'Dowd, C, Helm, M, Flasz, J, Arkoudis, E, Dossang, A, Hughes, C, Cassidy, E, McClelland, K, Odrzywol, E, Page, N, Barker, O, Miel, H. | | 登録日 | 2017-02-27 | | 公開日 | 2017-12-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Discovery and characterization of highly potent and selective allosteric USP7 inhibitors.
Nat. Chem. Biol., 14, 2018
|
|
8RIW
 
 | | T2R-TTL-1-L01 complex | | 分子名称: | (2-methyl-1~{H}-indol-5-yl) 3,4,5-trimethoxybenzenesulfonate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Prota, A.E.P, Boiarska, Z, Homer, J.A, Steinmetz, M.O, Moses, J.E. | | 登録日 | 2023-12-19 | | 公開日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | Modular synthesis of functional libraries by accelerated SuFEx click chemistry.
Chem Sci, 15, 2024
|
|
8U48
 
 | | Crystal structure of Bacteroides thetaiotamicron BT1285 D161A-E163A inactive Endoglycosidase in complex with high-mannose N-glycan (Man9GlcNAc2) substrate | | 分子名称: | Endo-beta-N-acetylglucosaminidase, PHOSPHATE ION, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | 著者 | Sastre, D.E, Sultana, N, Navarro, M.V.A.S, Sundberg, E.J. | | 登録日 | 2023-09-09 | | 公開日 | 2024-05-29 | | 最終更新日 | 2024-06-26 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Human gut microbes express functionally distinct endoglycosidases to metabolize the same N-glycan substrate.
Nat Commun, 15, 2024
|
|
8RIV
 
 | | T2R-TTL-1-K08 complex | | 分子名称: | (4-fluoranyl-2-methyl-1~{H}-indol-5-yl) 3,4,5-trimethoxybenzenesulfonate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | 著者 | Boiarska, Z, Homer, J.A, Steinmetz, M.O, Moses, J.E, Prota, A.E.P. | | 登録日 | 2023-12-19 | | 公開日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.78 Å) | | 主引用文献 | Modular synthesis of functional libraries by accelerated SuFEx click chemistry.
Chem Sci, 15, 2024
|
|
