8FCR
 
 | | Cryo-EM structure of p97:UBXD1 H4-bound state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | 著者 | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | 登録日 | 2022-12-01 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (4.12 Å) | | 主引用文献 | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8D81
 
 | | Cereblon~DDB1 bound to Pomalidomide | | 分子名称: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | 著者 | Watson, E.R, Lander, G.C. | | 登録日 | 2022-06-07 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
8FCQ
 
 | | Cryo-EM structure of p97:UBXD1 PUB-in state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | 著者 | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | 登録日 | 2022-12-01 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.93 Å) | | 主引用文献 | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8FCM
 
 | | Cryo-EM structure of p97:UBXD1 open state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase, UBX domain-containing protein 6 | | 著者 | Braxton, J.R, Tucker, M.R, Tse, E, Southworth, D.R. | | 登録日 | 2022-12-01 | | 公開日 | 2023-06-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.27 Å) | | 主引用文献 | The p97/VCP adaptor UBXD1 drives AAA+ remodeling and ring opening through multi-domain tethered interactions.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6PRU
 
 | | Photoconvertible crystals of PixJ from Thermosynechococcus elongatus | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Burgie, E.S, Clinger, J.A, Miller, M.D, Phillips Jr, G.N, Vierstra, R.D. | | 登録日 | 2019-07-11 | | 公開日 | 2019-12-18 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.539 Å) | | 主引用文献 | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8D7W
 
 | |
8CVP
 
 | | Cereblon-DDB1 in the Apo form | | 分子名称: | DNA damage-binding protein 1, Protein cereblon, ZINC ION | | 著者 | Watson, E.R, Lander, G.C. | | 登録日 | 2022-05-18 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
8D7V
 
 | |
8G57
 
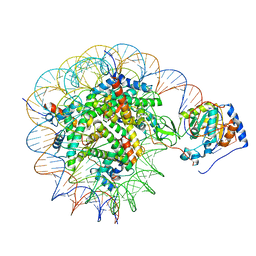 | | Structure of nucleosome-bound Sirtuin 6 deacetylase | | 分子名称: | DNA strand 1, DNA strand 2, Histone H2A type 1-B/E, ... | | 著者 | Chio, U.S, Rechiche, O, Bryll, A.R, Zhu, J, Feldman, J.L, Peterson, C.L, Tan, S, Armache, J.-P. | | 登録日 | 2023-02-11 | | 公開日 | 2023-04-26 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Cryo-EM structure of the human Sirtuin 6-nucleosome complex.
Sci Adv, 9, 2023
|
|
4Y81
 
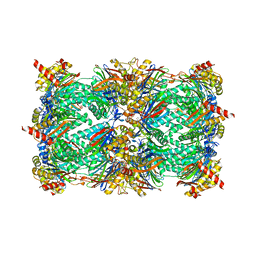 | | Yeast 20S proteasome in complex with Ac-PAY-ep | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-PAY-ep, CHLORIDE ION, ... | | 著者 | Huber, E.M, Groll, M. | | 登録日 | 2015-02-16 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
8QZ3
 
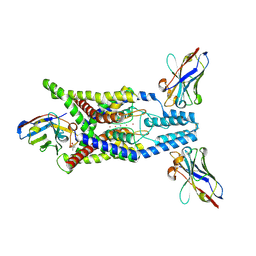 | | Crystal structure of human two pore domain potassium ion channel TREK-2 (K2P10.1) in complex with an activatory nanobody (Nb67) | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, Nanobody 67, POTASSIUM ION, ... | | 著者 | Baronina, A, Pike, A.C.W, Rodstrom, K.E.J, Ang, J, Bushell, S.R, Chalk, R, Mukhopadhyay, S.M.M, Pardon, E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N.A, Tucker, S.J, Steyaert, J, Carpenter, E.P, Structural Genomics Consortium (SGC) | | 登録日 | 2023-10-26 | | 公開日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Extracellular modulation of TREK-2 activity with nanobodies provides insight into the mechanisms of K2P channel regulation.
Nat Commun, 15, 2024
|
|
4Y8H
 
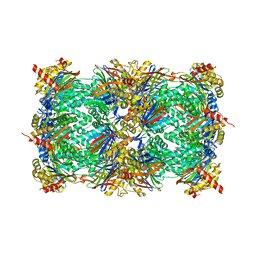 | | Yeast 20S proteasome in complex with N3-APAL-ep | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | 著者 | Huber, E.M, Groll, M. | | 登録日 | 2015-02-16 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
6FQM
 
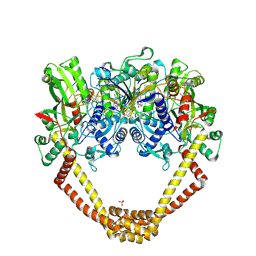 | | 3.06A COMPLEX OF S.AUREUS GYRASE with imidazopyrazinone T1 AND DNA | | 分子名称: | 7-[(3~{S})-3-azanylpyrrolidin-1-yl]-5-cyclopropyl-8-fluoranyl-imidazo[1,2-a]quinoxalin-4-one, DNA (5'-D(*GP*AP*GP*AP*GP*TP*AP*T*GP*GP*CP*CP*AP*TP*AP*CP*TP*CP*T)-3'), DNA gyrase subunit A, ... | | 著者 | Bax, B.D, Germe, T, Basque, E, Maxwell, A. | | 登録日 | 2018-02-14 | | 公開日 | 2018-04-04 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.06 Å) | | 主引用文献 | A new class of antibacterials, the imidazopyrazinones, reveal structural transitions involved in DNA gyrase poisoning and mechanisms of resistance.
Nucleic Acids Res., 46, 2018
|
|
6T21
 
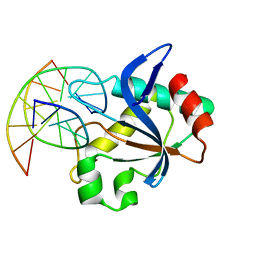 | | N-terminal domain of EcoKMcrA restriction endonuclease (NEco) in complex with T5mCGA target sequence | | 分子名称: | 5-methylcytosine-specific restriction enzyme A, DNA (5'-D(*GP*AP*AP*TP*(5CM)P*GP*AP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*TP*(5CM)P*GP*AP*TP*TP*C)-3') | | 著者 | Slyvka, A, Zagorskaite, E, Czapinska, H, Sasnauskas, G, Bochtler, M. | | 登録日 | 2019-10-07 | | 公開日 | 2019-10-23 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Crystal structure of the EcoKMcrA N-terminal domain (NEco): recognition of modified cytosine bases without flipping.
Nucleic Acids Res., 47, 2019
|
|
4Y8J
 
 | | Yeast 20S proteasome in complex with Ac-LLL-ep | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-PLL-ep, CHLORIDE ION, ... | | 著者 | Huber, E.M, Groll, M. | | 登録日 | 2015-02-16 | | 公開日 | 2015-06-17 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
6NB1
 
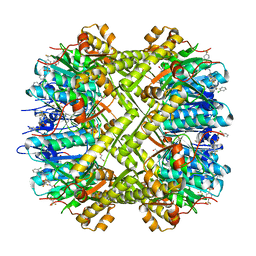 | | Crystal structure of Escherichia coli ClpP protease complexed with small molecule activator, ACP1-06 | | 分子名称: | ATP-dependent Clp protease proteolytic subunit, GLYCEROL, N-{2-[(2-chlorophenyl)sulfanyl]ethyl}-2-methyl-2-{[5-(trifluoromethyl)pyridin-2-yl]sulfonyl}propanamide | | 著者 | Mabanglo, M.F, Houry, W.A, Eger, B.T, Bryson, S, Pai, E.F. | | 登録日 | 2018-12-06 | | 公開日 | 2019-11-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | ClpP protease activation results from the reorganization of the electrostatic interaction networks at the entrance pores.
Commun Biol, 2, 2019
|
|
6UXO
 
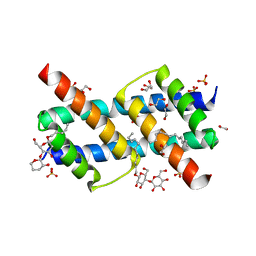 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with DDM | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Bcl-2 homologous antagonist/killer, ... | | 著者 | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | 登録日 | 2019-11-07 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.799 Å) | | 主引用文献 | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6PXV
 
 | | Cryo-EM structure of full-length insulin receptor bound to 4 insulin. 3D refinement was focused on the extracellular region. | | 分子名称: | Insulin, Insulin receptor | | 著者 | Uchikawa, E, Choi, E, Shang, G.J, Yu, H.T, Bai, X.C. | | 登録日 | 2019-07-27 | | 公開日 | 2019-09-04 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Activation mechanism of the insulin receptor revealed by cryo-EM structure of the fully liganded receptor-ligand complex.
Elife, 8, 2019
|
|
8GJH
 
 | | Salmonella ArnA | | 分子名称: | Bifunctional polymyxin resistance protein ArnA, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | 著者 | Sousa, M.C, Mitchell, M.E, Gatzeva-Topalova, P.Z. | | 登録日 | 2023-03-15 | | 公開日 | 2023-08-02 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (3.6 Å) | | 主引用文献 | Targeting the Conformational Change in ArnA Dehydrogenase for Selective Inhibition of Polymyxin Resistance.
Biochemistry, 62, 2023
|
|
7QD1
 
 | | Structure of the orange carotenoid protein from Planktothrix agardhii binding echinenone in the P21 space group | | 分子名称: | Orange carotenoid-binding protein, beta,beta-caroten-4-one | | 著者 | Andreeva, E.A, Hartmann, E, Schlichting, I, Colletier, J.-P. | | 登録日 | 2021-11-26 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Structure-function-dynamics relationships in the peculiar Planktothrix PCC7805 OCP1: Impact of his-tagging and carotenoid type.
Biochim Biophys Acta Bioenerg, 1863, 2022
|
|
7QD0
 
 | | Structure of the orange carotenoid protein from Planktothrix agardhii binding echinenone in the C2 space group | | 分子名称: | ACETATE ION, ARGININE, GLYCEROL, ... | | 著者 | Andreeva, E.A, Hartmann, E, Schlichting, I, Colletier, J.-P. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-function-dynamics relationships in the peculiar Planktothrix PCC7805 OCP1: Impact of his-tagging and carotenoid type.
Biochim Biophys Acta Bioenerg, 1863, 2022
|
|
7QD2
 
 | | Structure of the orange carotenoid protein from Planktothrix agardhii binding canthaxanthin in the P21 space group | | 分子名称: | ACETATE ION, GLYCEROL, Orange carotenoid-binding protein, ... | | 著者 | Andreeva, E.A, Hartmann, E, Schlichting, I, Colletier, J.-P. | | 登録日 | 2021-11-26 | | 公開日 | 2022-07-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Structure-function-dynamics relationships in the peculiar Planktothrix PCC7805 OCP1: Impact of his-tagging and carotenoid type.
Biochim Biophys Acta Bioenerg, 1863, 2022
|
|
7OJK
 
 | | Lassa virus L protein bound to the distal promoter duplex [DISTAL-PROMOTER] | | 分子名称: | 3' RNA, 5' RNA, RNA-directed RNA polymerase L, ... | | 著者 | Kouba, T, Vogel, D, Thorkelsson, S, Quemin, E, Williams, H.M, Milewski, M, Busch, C, Gunther, S, Grunewald, K, Rosenthal, M, Cusack, S. | | 登録日 | 2021-05-16 | | 公開日 | 2021-12-01 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (3.89 Å) | | 主引用文献 | Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Nat Commun, 12, 2021
|
|
5N0A
 
 | | Crystal structure of A259C covalently linked dengue 2 virus envelope glycoprotein dimer in complex with the Fab fragment of the broadly neutralizing human antibody EDE2 A11 | | 分子名称: | BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 HEAVY CHAIN, Envelope Glycoprotein E, ... | | 著者 | Vaney, M.C, Rouvinski, A, Guardado-Calvo, P, Sharma, A, Rey, F.A. | | 登録日 | 2017-02-02 | | 公開日 | 2017-06-07 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | Covalently linked dengue virus envelope glycoprotein dimers reduce exposure of the immunodominant fusion loop epitope.
Nat Commun, 8, 2017
|
|
7QEI
 
 | | Structure of human MTHFD2L in complex with TH7299 | | 分子名称: | (2S)-2-[[4-[[2,4-bis(azanyl)-6-oxidanylidene-1H-pyrimidin-5-yl]carbamoylamino]phenyl]carbonylamino]pentanedioic acid, 2'-MONOPHOSPHOADENOSINE-5'-DIPHOSPHATE, Probable bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase 2 | | 著者 | Gustafsson, R, Scaletti, E.R, Stenmark, P. | | 登録日 | 2021-12-03 | | 公開日 | 2022-10-12 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The First Structure of Human MTHFD2L and Its Implications for the Development of Isoform-Selective Inhibitors.
Chemmedchem, 17, 2022
|
|
