5I4A
 
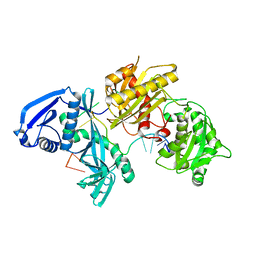 | | X-ray crystal structure of Marinitoga piezophila Argonaute in complex with 5' OH guide RNA | | 分子名称: | Argonaute protein, RNA (5'-R(*UP*AP*UP*AP*CP*AP*AP*CP*CP*UP*AP*CP*UP*U)-3') | | 著者 | Doxzen, K.W, Kaya, E, Knoll, K.R, Wilson, R.C, Strutt, S.C, Kranzusch, P.J, Doudna, J.A. | | 登録日 | 2016-02-11 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.949 Å) | | 主引用文献 | A bacterial Argonaute with noncanonical guide RNA specificity.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8HUV
 
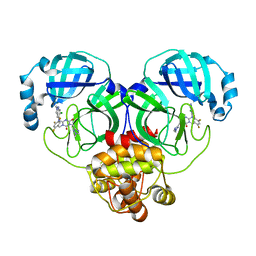 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with S217622 | | 分子名称: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-[(1-methyl-1,2,4-triazol-3-yl)methyl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | 著者 | Zeng, P, Zhang, J, Li, J. | | 登録日 | 2022-12-24 | | 公開日 | 2023-06-21 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Structural basis for the inhibition of coronaviral main proteases by ensitrelvir.
Structure, 31, 2023
|
|
4LI5
 
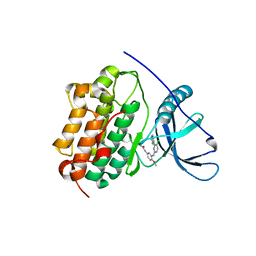 | | EGFR-K IN COMPLEX WITH N-[3-[[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino]-4-methoxy-phenyl] Prop-2-enamide | | 分子名称: | Epidermal growth factor receptor, N-(3-{[5-chloro-4-(1H-indol-3-yl)pyrimidin-2-yl]amino}-4-methoxyphenyl)propanamide, SODIUM ION | | 著者 | Debreczeni, J.E, Seiffert, G.B, Kiefersauer, R, Augustin, M, Nagel, S, Ward, R, Anderton, M, Ashton, S, Bethel, P, Box, M, Butterworth, S, Colclough, N, Chroley, C, Chuaqui, C, Cross, D, Eberlein, C, Finlay, R, Hill, G, Grist, M, Klinowska, T, Lane, C, Martin, S, Orme, J, Smith, P, Wang, F, Waring, M. | | 登録日 | 2013-07-02 | | 公開日 | 2013-08-28 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | Structure- and Reactivity-Based Development of Covalent Inhibitors of the Activating and Gatekeeper Mutant Forms of the Epidermal Growth Factor Receptor (EGFR).
J.Med.Chem., 56, 2013
|
|
2YFN
 
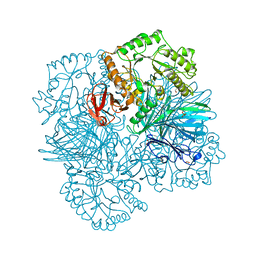 | | galactosidase domain of alpha-galactosidase-sucrose kinase, AgaSK | | 分子名称: | 1,2-ETHANEDIOL, ALPHA-GALACTOSIDASE-SUCROSE KINASE AGASK, GLYCEROL, ... | | 著者 | Sulzenbacher, G, Bruel, L, Tison-Cervera, M, Pujol, A, Nicoletti, C, Perrier, J, Galinier, A, Ropartz, D, Fons, M, Pompeo, F, Giardina, T. | | 登録日 | 2011-04-07 | | 公開日 | 2011-09-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Agask, a Bifunctional Enzyme from the Human Microbiome Coupling Galactosidase and Kinase Activities
J.Biol.Chem., 286, 2011
|
|
4C7W
 
 | |
5HSZ
 
 | |
8I30
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with 32j | | 分子名称: | (2~{R})-1-[4,4-bis(fluoranyl)cyclohexyl]carbonyl-4,4-bis(fluoranyl)-~{N}-[(2~{R},3~{S})-3-oxidanyl-4-oxidanylidene-1-phenyl-4-(pyridin-2-ylmethylamino)butan-2-yl]pyrrolidine-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | 著者 | Zeng, R, Huang, C, Xie, L.W, Wang, K, Liu, Y.Z, Yang, S.Y, Lei, J. | | 登録日 | 2023-01-16 | | 公開日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Discovery and structure-activity relationship studies of novel alpha-ketoamide derivatives targeting the SARS-CoV-2 main protease.
Eur.J.Med.Chem., 259, 2023
|
|
3AQO
 
 | |
8JOP
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with 11a | | 分子名称: | 3C-like proteinase nsp5, methyl (6~{R})-5-ethanoyl-7-oxidanylidene-6-[4-(trifluoromethyl)phenyl]-8,9,10,11-tetrahydro-6~{H}-benzo[b][1,4]benzodiazepine-2-carboxylate | | 著者 | Zeng, R, Liu, Y.Z, Wang, F.L, Yang, S.Y, Lei, J. | | 登録日 | 2023-06-08 | | 公開日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Discovery of benzodiazepine derivatives as a new class of covalent inhibitors of SARS-CoV-2 main protease.
Bioorg.Med.Chem.Lett., 92, 2023
|
|
4C7L
 
 | | Crystal structure of Mouse Hepatitis virus strain S Hemagglutinin- esterase | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | 著者 | Zeng, Q.H, Huizinga, E.G. | | 登録日 | 2013-09-23 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Murine Coronavirus Hemagglutinin-Esterase Receptor-Binding Site: A Major Shift in Ligand Specificity Through Modest Changes in Architecture.
Plos Pathog., 8, 2012
|
|
2ND0
 
 | |
2NCZ
 
 | |
2NDF
 
 | |
2NDG
 
 | |
2ND1
 
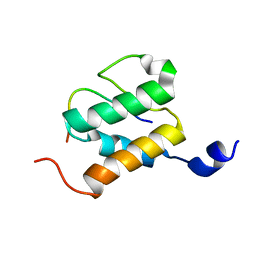 | |
2IXA
 
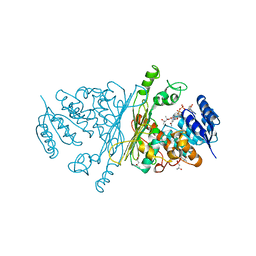 | | A-zyme, N-acetylgalactosaminidase | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ALPHA-N-ACETYLGALACTOSAMINIDASE, ... | | 著者 | Sulzenbacher, G, Liu, Q.P, Bourne, Y, Henrissat, B, Clausen, H. | | 登録日 | 2006-07-07 | | 公開日 | 2007-04-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Bacterial Glycosidases for the Production of Universal Red Blood Cells.
Nat.Biotechnol., 25, 2007
|
|
2IVX
 
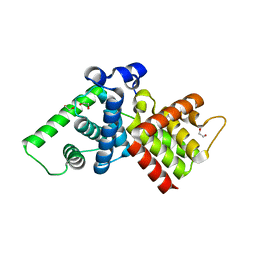 | | Crystal structure of human cyclin T2 at 1.8 A resolution | | 分子名称: | 1,2-ETHANEDIOL, CYCLIN-T2 | | 著者 | Debreczeni, J.E, Bullock, A.N, Fedorov, O, Savitsky, P, Berridge, G, Das, S, Pike, A.C.W, Turnbull, A, Ugochukwu, E, Papagrigoriou, E, Gorrec, F, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, von Delft, F, Knapp, S. | | 登録日 | 2006-06-21 | | 公開日 | 2006-07-19 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation.
EMBO J., 27, 2008
|
|
2IX9
 
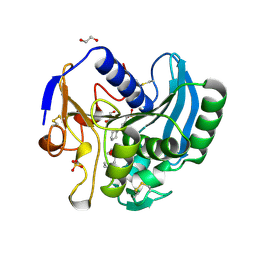 | |
2IZV
 
 | | CRYSTAL STRUCTURE OF SOCS-4 IN COMPLEX WITH ELONGIN-B AND ELONGIN-C AT 2.55A RESOLUTION | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, SODIUM ION, ... | | 著者 | Debreczeni, J.E, Bullock, A, Papagrigoriou, E, Turnbull, A, Pike, A.C.W, Gorrec, F, von Delft, F, Sundstrom, M, Arrowsmith, C, Weigelt, J, Edwards, A, Knapp, S. | | 登録日 | 2006-07-26 | | 公開日 | 2006-08-02 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structure of the SOCS4-ElonginB/C complex reveals a distinct SOCS box interface and the molecular basis for SOCS-dependent EGFR degradation.
Structure, 15, 2007
|
|
6IYX
 
 | | Crystal Structure Analysis of a Eukaryotic Membrane Protein | | 分子名称: | BROMIDE ION, CALCIUM ION, Trimeric intracellular cation channel type A | | 著者 | Zeng, Y, Wang, X.H, Gao, F, Su, M, Chen, Y.H. | | 登録日 | 2018-12-17 | | 公開日 | 2019-05-01 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for activity of TRIC counter-ion channels in calcium release.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6IZ3
 
 | |
2IXB
 
 | | Crystal structure of N-ACETYLGALACTOSAMINIDASE in complex with GalNAC | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, ... | | 著者 | Sulzenbacher, G, Liu, Q.P, Bourne, Y, Henrissat, B, Clausen, H. | | 登録日 | 2006-07-07 | | 公開日 | 2007-04-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Bacterial Glycosidases for the Production of Universal Red Blood Cells.
Nat.Biotechnol., 25, 2007
|
|
2LIX
 
 | |
4IBQ
 
 | | Human p53 core domain with hot spot mutation R273C | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Cellular tumor antigen p53, ... | | 著者 | Rozenberg, H, Eldar, A, Diskin-Posner, Y, Shakked, Z. | | 登録日 | 2012-12-09 | | 公開日 | 2013-08-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural studies of p53 inactivation by DNA-contact mutations and its rescue by suppressor mutations via alternative protein-DNA interactions.
Nucleic Acids Res., 41, 2013
|
|
4IBS
 
 | | Human p53 core domain with hot spot mutation R273H (form I) | | 分子名称: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, ZINC ION | | 著者 | Rozenberg, H, Eldar, A, Diskin-Posner, Y, Shakked, Z. | | 登録日 | 2012-12-09 | | 公開日 | 2013-08-14 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Structural studies of p53 inactivation by DNA-contact mutations and its rescue by suppressor mutations via alternative protein-DNA interactions.
Nucleic Acids Res., 41, 2013
|
|
