1EC3
 
 | | HIV-1 protease in complex with the inhibitor MSA367 | | 分子名称: | HIV-1 PROTEASE, N,N-[2,5-O-DIBENZYL-GLUCARYL]-DI-[VALINYL-AMINOMETHANYL-PYRIDINE] | | 著者 | Unge, T. | | 登録日 | 2000-01-25 | | 公開日 | 2002-06-26 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
3C59
 
 | |
1EV9
 
 | | RAT GLUTATHIONE S-TRANSFERASE A1-1 MUTANT W21F WITH GSO3 BOUND | | 分子名称: | GLUTATHIONE S-TRANSFERASE A1-1, GLUTATHIONE SULFONIC ACID, SULFATE ION | | 著者 | Adman, E.T, Le Trong, I, Stenkamp, R.E, Nieslanik, B.S, Dietze, E.C, Tai, G, Ibarra, C, Atkins, W.M. | | 登録日 | 2000-04-19 | | 公開日 | 2001-02-07 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Localization of the C-terminus of rat glutathione S-transferase A1-1: crystal structure of mutants W21F and W21F/F220Y.
Proteins, 42, 2001
|
|
4RW7
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (K103N, Y181C) variant in complex with (E)-3-(3-chloro-5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)phenyl)acrylonitrile (JLJ532), a non-nucleoside inhibitor | | 分子名称: | (2E)-3-(3-chloro-5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}phenyl)prop-2-enenitrile, Reverse transcriptase/ribonuclease H, p51 subunit, ... | | 著者 | Frey, K.M, Anderson, K.S. | | 登録日 | 2014-12-01 | | 公開日 | 2015-04-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.014 Å) | | 主引用文献 | Structure-Based Evaluation of Non-nucleoside Inhibitors with Improved Potency and Solubility That Target HIV Reverse Transcriptase Variants.
J.Med.Chem., 58, 2015
|
|
1EWD
 
 | |
1E2I
 
 | | The nucleoside binding site of Herpes simplex type 1 thymidine kinase analyzed by X-ray crystallography | | 分子名称: | 9-HYDROXYPROPYLADENINE, R-ISOMER, S-ISOMER, ... | | 著者 | Vogt, J, Scapozza, L, Schulz, G.E. | | 登録日 | 2000-05-23 | | 公開日 | 2000-11-06 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Nucleoside Binding Site of Herpes Simplex Type 1 Thymidine Kinase Analyzed by X-Ray Crystallography
Proteins, 41, 2000
|
|
4RW4
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (K103N,Y181C) variant in complex with (E)-3-(3-chloro-5-(4-chloro-2-(2-(2,4-dioxo-3,4- dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)phenyl)acrylonitrile (JLJ494), a Non-nucleoside Inhibitor | | 分子名称: | (2E)-3-(3-chloro-5-{4-chloro-2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}phenyl)prop-2-enenitrile, Reverse transcriptase/ribonuclease H, p51 subunit, ... | | 著者 | Frey, K.M, Anderson, K.S. | | 登録日 | 2014-12-01 | | 公開日 | 2015-04-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.674 Å) | | 主引用文献 | Structure-Based Evaluation of Non-nucleoside Inhibitors with Improved Potency and Solubility That Target HIV Reverse Transcriptase Variants.
J.Med.Chem., 58, 2015
|
|
1D07
 
 | | Hydrolytic haloalkane dehalogenase linb from sphingomonas paucimobilis UT26 with 1,3-propanediol, a product of debromidation of dibrompropane, at 2.0A resolution | | 分子名称: | 1,3-PROPANDIOL, BROMIDE ION, HALOALKANE DEHALOGENASE | | 著者 | Marek, J, Vevodova, J, Damborsky, J, Smatanova, I, Svensson, L.A, Newman, J, Nagata, Y, Takagi, M. | | 登録日 | 1999-09-09 | | 公開日 | 2000-09-11 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the haloalkane dehalogenase from Sphingomonas paucimobilis UT26.
Biochemistry, 39, 2000
|
|
4RVJ
 
 | | Crystal structure of multidrug-resistant clinical isolate A02 HIV-1 protease in complex with amprenavir | | 分子名称: | HIV-1 protease, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | 著者 | Yedidi, R.S, Garimella, H, Kaufman, J.D, Das, D, Wingfield, P.T, Mitsuya, H. | | 登録日 | 2014-11-26 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Enhanced antiviral activity by the P2-tris-tetrahydrofuran moiety of GRL-0519, a novel nonpeptidic HIV-1 protease inhibitor (PI), against multi-PI-resistant and highly darunavir-resistant strains of HIV-1
To be Published
|
|
4RW8
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in complex with (E)-3-(3-chloro-5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)phenyl)acrylonitrile (JLJ532), a non-nucleoside inhibitor' | | 分子名称: | (2E)-3-(3-chloro-5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}phenyl)prop-2-enenitrile, Reverse transcriptase/ribonuclease H, p51 subunit, ... | | 著者 | Frey, K.M, Anderson, K.S. | | 登録日 | 2014-12-01 | | 公開日 | 2015-04-29 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.878 Å) | | 主引用文献 | Structure-Based Evaluation of Non-nucleoside Inhibitors with Improved Potency and Solubility That Target HIV Reverse Transcriptase Variants.
J.Med.Chem., 58, 2015
|
|
2GMV
 
 | | PEPCK complex with a GTP-competitive inhibitor | | 分子名称: | MANGANESE (II) ION, N-(4-{[3-BUTYL-1-(2-FLUOROBENZYL)-2,6-DIOXO-2,3,6,7-TETRAHYDRO-1H-PURIN-8-YL]METHYL}PHENYL)-1-METHYL-1H-IMIDAZOLE-4-SULFONAMIDE, PHOSPHOENOLPYRUVATE, ... | | 著者 | Dunten, P. | | 登録日 | 2006-04-07 | | 公開日 | 2007-05-29 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | C-8 Modifications of 3-alkyl-1,8-dibenzylxanthines as inhibitors of human cytosolic phosphoenolpyruvate carboxykinase.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4RL2
 
 | | Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins | | 分子名称: | (1R)-2-({(R)-carboxy[(2R,5S)-4-carboxy-5-methyl-5,6-dihydro-2H-1,3-thiazin-2-yl]methyl}amino)-2-oxo-1-phenylethanaminium, Beta-lactamase NDM-1, ZINC ION | | 著者 | Feng, H, Ding, J, Zhu, D, Liu, X, Xu, X, Zhang, Y, Zang, S, Wang, D.-C, Liu, W. | | 登録日 | 2014-10-15 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.008 Å) | | 主引用文献 | Structural and Mechanistic Insights into NDM-1 Catalyzed Hydrolysis of Cephalosporins.
J.Am.Chem.Soc., 136, 2014
|
|
1ISP
 
 | | Crystal structure of Bacillus subtilis lipase at 1.3A resolution | | 分子名称: | GLYCEROL, lipase | | 著者 | Kawasaki, K, Kondo, H, Suzuki, M, Ohgiya, S, Tsuda, S. | | 登録日 | 2001-12-19 | | 公開日 | 2002-12-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Alternate conformations observed in catalytic serine of Bacillus subtilis lipase determined at 1.3 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1E2H
 
 | | The nucleoside binding site of Herpes simplex type 1 thymidine kinase analyzed by X-ray crystallography | | 分子名称: | SULFATE ION, THYMIDINE KINASE | | 著者 | Vogt, J, Scapozza, L, Schulz, G.E. | | 登録日 | 2000-05-23 | | 公開日 | 2000-11-06 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Nucleoside Binding Site of Herpes Simplex Type 1 Thymidine Kinase Analyzed by X-Ray Crystallography
Proteins: Struct.,Funct., Genet., 41, 2000
|
|
1VPB
 
 | |
4CSA
 
 | | Crystal structure of the asymmetric human metapneumovirus M2-1 tetramer bound to a DNA 4-mer | | 分子名称: | 5'-D(*AP*GP*TP*TP*AP)-3', GLYCEROL, M2-1, ... | | 著者 | Leyrat, C, Renner, M, Harlos, K, Grimes, J.M. | | 登録日 | 2014-03-05 | | 公開日 | 2014-05-28 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.28 Å) | | 主引用文献 | Drastic Changes in Conformational Dynamics of the Antiterminator M2-1 Regulate Transcription Efficiency in Pneumovirinae.
Elife, 3, 2014
|
|
3AO4
 
 | | Fragment-based approach to the design of ligands targeting a novel site on HIV-1 integrase | | 分子名称: | 3-(1,3-benzodioxol-5-yl)-1-methyl-1H-pyrazol-5-amine, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Wielens, J, Headey, S.J, Parker, M.W, Chalmers, D.K, Scanlon, M.J. | | 登録日 | 2010-09-20 | | 公開日 | 2011-03-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
3AO3
 
 | | Fragment-based approach to the design of ligands targeting a novel site on HIV-1 integrase | | 分子名称: | 3-(1,3-benzodioxol-5-yl)-1-methyl-1H-pyrazole-5-carboxylic acid, CADMIUM ION, CHLORIDE ION, ... | | 著者 | Wielens, J, Headey, S.J, Parker, M.W, Chalmers, D.K, Scanlon, M.J. | | 登録日 | 2010-09-20 | | 公開日 | 2011-03-09 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Fragment-based design of ligands targeting a novel site on the integrase enzyme of human immunodeficiency virus 1
Chemmedchem, 6, 2011
|
|
3F5E
 
 | | Crystal structure of Toxoplasma gondii micronemal protein 1 bound to 2'F-3'SiaLacNAc1-3 | | 分子名称: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | 著者 | Garnett, J.A, Liu, Y, Feizi, T, Matthews, S.J. | | 登録日 | 2008-11-03 | | 公開日 | 2009-07-28 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Detailed insights from microarray and crystallographic studies into carbohydrate recognition by microneme protein 1 (MIC1) of Toxoplasma gondii.
Protein Sci., 18, 2009
|
|
4K00
 
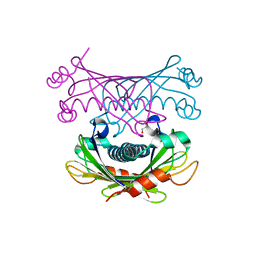 | | Crystal structure of Slr0204, a 1,4-dihydroxy-2-naphthoyl-CoA thioesterase from Synechocystis | | 分子名称: | 1,2-ETHANEDIOL, 1,4-dihydroxy-2-naphthoyl-CoA hydrolase | | 著者 | Furt, F, Allen, W.J, Widhalm, J.R, Madzelan, P, Rizzo, R.C, Basset, G, Wilson, M.A. | | 登録日 | 2013-04-03 | | 公開日 | 2013-04-17 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Functional convergence of structurally distinct thioesterases from cyanobacteria and plants involved in phylloquinone biosynthesis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2JLC
 
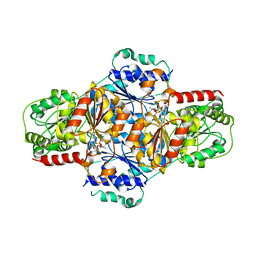 | | Crystal structure of E.coli MenD, 2-succinyl-5-enolpyruvyl-6-hydroxy- 3-cyclohexadiene-1-carboxylate synthase - native protein | | 分子名称: | 2-SUCCINYL-5-ENOLPYRUVYL-6-HYDROXY-3-CYCLOHEXENE -1-CARBOXYLATE SYNTHASE, MANGANESE (II) ION, THIAMINE DIPHOSPHATE | | 著者 | Dawson, A, Fyfe, P.K, Hunter, W.N. | | 登録日 | 2008-09-08 | | 公開日 | 2008-10-21 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Specificity and Reactivity in Menaquinone Biosynthesis: The Structure of Escherichia Coli Mend (2-Succinyl-5-Enolpyruvyl-6-Hydroxy-3-Cyclohexadiene-1-Carboxylate Synthase).
J.Mol.Biol., 384, 2008
|
|
1UKR
 
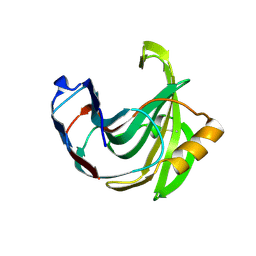 | | STRUCTURE OF ENDO-1,4-BETA-XYLANASE C | | 分子名称: | ENDO-1,4-B-XYLANASE I | | 著者 | Krengel, U, Dijkstra, B.W. | | 登録日 | 1996-08-23 | | 公開日 | 1997-12-24 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Three-dimensional structure of Endo-1,4-beta-xylanase I from Aspergillus niger: molecular basis for its low pH optimum.
J.Mol.Biol., 263, 1996
|
|
1DMP
 
 | | STRUCTURE OF HIV-1 PROTEASE COMPLEX | | 分子名称: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-1,3-BIS([(3-AMINO)PHENYL]METHYL)-4,7-BIS(PHENYLMETHYL)-2H-1,3-DIAZEPINONE | | 著者 | Chang, C.-H. | | 登録日 | 1996-11-01 | | 公開日 | 1997-11-12 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Improved cyclic urea inhibitors of the HIV-1 protease: synthesis, potency, resistance profile, human pharmacokinetics and X-ray crystal structure of DMP 450.
Chem.Biol., 3, 1996
|
|
1DD6
 
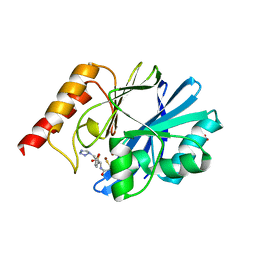 | | IMP-1 METALLO BETA-LACTAMASE FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH A MERCAPTOCARBOXYLATE INHIBITOR | | 分子名称: | (2-MERCAPTOMETHYL-4-PHENYL-BUTYRYLIMINO)-(5-TETRAZOL-1-YLMETHYL-THIOPHEN-2-YL)-ACETIC ACID, IMP-1 METALLO BETA-LACTAMASE, SULFATE ION, ... | | 著者 | Concha, N.O, Janson, C.A, Rowling, P, Pearson, S, Cheever, C.A, Clarke, B.P, Lewis, C, Galleni, M, Frere, J.M, Payne, D.J, Bateson, J.H, Abdel-Meguid, S.S. | | 登録日 | 1999-11-08 | | 公開日 | 2000-11-08 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal structure of the IMP-1 metallo beta-lactamase from Pseudomonas aeruginosa and its complex with a mercaptocarboxylate inhibitor: binding determinants of a potent, broad-spectrum inhibitor.
Biochemistry, 39, 2000
|
|
4IED
 
 | | Crystal Structure of FUS-1 (OXA-85), a Class D beta-lactamase from Fusobacterium nucleatum subsp. polymorphum | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Class D beta-lactamase, ... | | 著者 | Mangani, S, Benvenuti, M, Docquier, J.D. | | 登録日 | 2012-12-13 | | 公開日 | 2014-01-22 | | 最終更新日 | 2025-03-26 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal Structure of FUS-1 (OXA-85), a Class D beta-lactamase from Fusobacterium nucleatum subsp. polymorphum
To be Published
|
|
