6MSB
 
 | |
7M2U
 
 | |
6MSK
 
 | |
8P7L
 
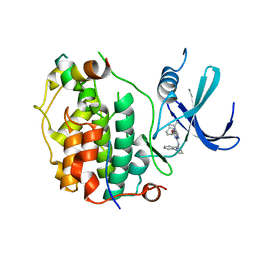 | | Cryo-EM structure of CDK7 subunit of CAK in complex with inhibitor LDC4297 | | 分子名称: | 2-[(3S)-piperidin-3-yl]oxy-8-propan-2-yl-N-[(2-pyrazol-1-ylphenyl)methyl]pyrazolo[1,5-a][1,3,5]triazin-4-amine, CDK-activating kinase assembly factor MAT1, Cyclin-dependent kinase 7 | | 著者 | Cushing, V.I, Koh, A.F, Feng, J, Jurgaityte, K, Bahl, A.K, Ali, S, Kotecha, A, Greber, B.J. | | 登録日 | 2023-05-30 | | 公開日 | 2024-03-20 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (2.1 Å) | | 主引用文献 | High-resolution cryo-EM of the human CDK-activating kinase for structure-based drug design.
Nat Commun, 15, 2024
|
|
7UXH
 
 | |
7UXC
 
 | |
6MSD
 
 | |
6MSE
 
 | |
6MSH
 
 | |
8VNZ
 
 | |
6V92
 
 | | RSC-NCP | | 分子名称: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC14, ... | | 著者 | Patel, A.B, Moore, C.M, Greber, B.J, Nogales, E. | | 登録日 | 2019-12-13 | | 公開日 | 2020-01-15 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (20 Å) | | 主引用文献 | Architecture of the chromatin remodeler RSC and insights into its nucleosome engagement.
Elife, 8, 2019
|
|
5LN3
 
 | | The human 26S Proteasome at 6.8 Ang. | | 分子名称: | 26S protease regulatory subunit 10B, 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, ... | | 著者 | Schweitzer, A, Beck, F, Sakata, E, Unverdorben, P. | | 登録日 | 2016-08-03 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (6.8 Å) | | 主引用文献 | Molecular Details Underlying Dynamic Structures and Regulation of the Human 26S Proteasome.
Mol. Cell Proteomics, 16, 2017
|
|
7KW7
 
 | | Atomic cryoEM structure of Hsp90-Hsp70-Hop-GR | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Glucocorticoid receptor, Heat shock 70 kDa protein 1A, ... | | 著者 | Wang, R.Y, Noddings, C.M, Kirschke, E, Myasnikov, A, Johnson, J.L, Agard, D.A. | | 登録日 | 2020-11-30 | | 公開日 | 2021-12-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.57 Å) | | 主引用文献 | Structure of Hsp90-Hsp70-Hop-GR reveals the Hsp90 client-loading mechanism.
Nature, 601, 2022
|
|
7RPW
 
 | |
7RPO
 
 | |
7RPX
 
 | |
8PQL
 
 | | K48-linked ubiquitin chain formation with a cullin-RING E3 ligase and Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2-donor UB-acceptor UB-SIL1 peptide | | 分子名称: | 5-azanylpentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | 登録日 | 2023-07-11 | | 公開日 | 2024-02-14 | | 最終更新日 | 2025-07-09 | | 実験手法 | ELECTRON MICROSCOPY (3.76 Å) | | 主引用文献 | Mechanism of millisecond Lys48-linked poly-ubiquitin chain formation by cullin-RING ligases.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OF4
 
 | | Nucleosome Bound human SIRT6 (Composite) | | 分子名称: | DNA (145-MER), Histone H2A type 1, Histone H2B, ... | | 著者 | Smirnova, E, Bignon, E, Schultz, P, Papai, G, Ben-Shem, A. | | 登録日 | 2023-03-13 | | 公開日 | 2023-08-09 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (2.94 Å) | | 主引用文献 | Binding to nucleosome poises human SIRT6 for histone H3 deacetylation.
Elife, 12, 2024
|
|
9GMK
 
 | | SIRT7:H3K18DTU nucleosome complex | | 分子名称: | DNA (148-MER), Histone H2A type 2-A, Histone H2B type 1-J, ... | | 著者 | Moreno-Yruela, C, Ekundayo, B, Foteva, P, Calvino-Sanles, E, Ni, D, Stahlberg, H, Fierz, B. | | 登録日 | 2024-08-29 | | 公開日 | 2025-01-29 | | 最終更新日 | 2025-07-09 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of SIRT7 nucleosome engagement and substrate specificity.
Nat Commun, 16, 2025
|
|
8Q7R
 
 | | Ubiquitin ligation to substrate by a cullin-RING E3 ligase & Cdc34: NEDD8-CUL2-RBX1-ELOB/C-FEM1C with trapped UBE2R2~donor UB-Sil1 peptide | | 分子名称: | 5-azanyl-1-oxidanyl-pentan-2-one, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | 著者 | Liwocha, J, Prabu, J.R, Kleiger, G, Schulman, B.A. | | 登録日 | 2023-08-16 | | 公開日 | 2024-02-21 | | 最終更新日 | 2025-07-09 | | 実験手法 | ELECTRON MICROSCOPY (3.71 Å) | | 主引用文献 | Cullin-RING ligases employ geometrically optimized catalytic partners for substrate targeting.
Mol.Cell, 84, 2024
|
|
6K1P
 
 | | The complex of ISWI-nucleosome in the ADP.BeF-bound state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | 著者 | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | 登録日 | 2019-05-10 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-09-25 | | 実験手法 | ELECTRON MICROSCOPY (3.87 Å) | | 主引用文献 | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6JYL
 
 | | The crosslinked complex of ISWI-nucleosome in the ADP.BeF-bound state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | 著者 | Yan, L.J, Wu, H, Li, X.M, Gao, N, Chen, Z.C. | | 登録日 | 2019-04-26 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Structures of the ISWI-nucleosome complex reveal a conserved mechanism of chromatin remodeling.
Nat.Struct.Mol.Biol., 26, 2019
|
|
7X3X
 
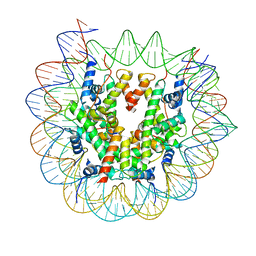 | | Cryo-EM structure of N1 nucleosome-RA | | 分子名称: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | 著者 | Lifei, L, Kangjing, C, Chen, Z. | | 登録日 | 2022-03-01 | | 公開日 | 2023-09-20 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7X3W
 
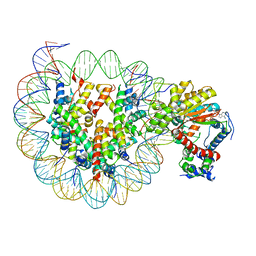 | | Cryo-EM structure of ISW1-N1 nucleosome | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (146-MER), ... | | 著者 | Lifei, L, Kangjing, C, Chen, Z. | | 登録日 | 2022-03-01 | | 公開日 | 2023-09-20 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure of the ISW1a complex bound to the dinucleosome.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4UVA
 
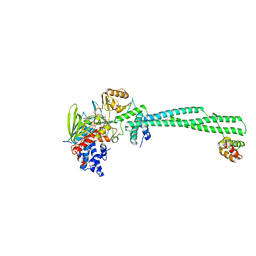 | | LSD1(KDM1A)-CoREST in complex with 1-Methyl-Tranylcypromine (1R,2S) | | 分子名称: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl (2R,3S,4S)-2,3,4-trihydroxy-5-[(4aS)-4a-[(1S,3E)-3-imino-1-phenylbutyl]-7,8-dimethyl-2,4-dioxo-3,4,4a,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]pentyl dihydrogen diphosphate | | 著者 | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | 登録日 | 2014-08-05 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
