2NXX
 
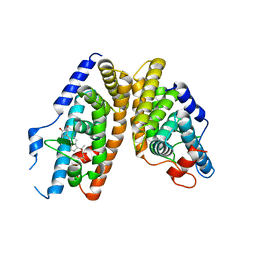 | | Crystal Structure of the Ligand-Binding Domains of the T.castaneum (Coleoptera) Heterodimer EcrUSP Bound to Ponasterone A | | 分子名称: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, Ecdysone Receptor (EcR, NRH1), ... | | 著者 | Iwema, T, Billas, I, Moras, D. | | 登録日 | 2006-11-20 | | 公開日 | 2007-10-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structural and functional characterization of a novel type of ligand-independent RXR-USP receptor.
Embo J., 26, 2007
|
|
2OEM
 
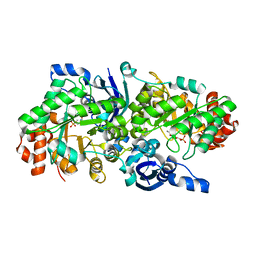 | | Crystal structure of a rubisco-like protein from Geobacillus kaustophilus liganded with Mg2+ and 2,3-diketohexane 1-phosphate | | 分子名称: | (1Z)-2-HYDROXY-3-OXOHEX-1-EN-1-YL DIHYDROGEN PHOSPHATE, 2,3-diketo-5-methylthiopentyl-1-phosphate enolase, MAGNESIUM ION | | 著者 | Fedorov, A.A, Imker, H.J, Fedorov, E.V, Gerlt, J.A, Almo, S.C. | | 登録日 | 2006-12-30 | | 公開日 | 2007-03-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mechanistic Diversity in the RuBisCO Superfamily: The "Enolase" in the Methionine Salvage Pathway in Geobacillus kaustophilus.
Biochemistry, 46, 2007
|
|
2OSX
 
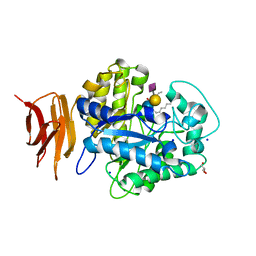 | | Endo-glycoceramidase II from Rhodococcus sp.: Ganglioside GM3 Complex | | 分子名称: | Endoglycoceramidase II, GLYCEROL, N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)PALMITAMIDE, ... | | 著者 | Caines, M.E.C, Strynadka, N.C.J. | | 登録日 | 2007-02-06 | | 公開日 | 2007-02-27 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Structural and Mechanistic Analyses of endo-Glycoceramidase II, a Membrane-associated Family 5 Glycosidase in the Apo and GM3 Ganglioside-bound Forms.
J.Biol.Chem., 282, 2007
|
|
2Q9F
 
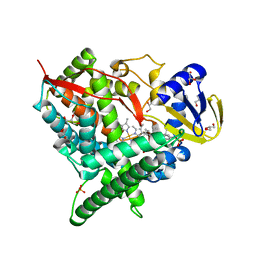 | | Crystal structure of human cytochrome P450 46A1 in complex with cholesterol-3-sulphate | | 分子名称: | CHOLEST-5-EN-3-YL HYDROGEN SULFATE, Cytochrome P450 46A1, GLYCEROL, ... | | 著者 | White, M.A, Mast, N.V, Johnson, E.F, Stout, C.D, Pikuleva, I.A. | | 登録日 | 2007-06-12 | | 公開日 | 2008-06-17 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of substrate-bound and substrate-free cytochrome P450 46A1, the principal cholesterol hydroxylase in the brain.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Q9M
 
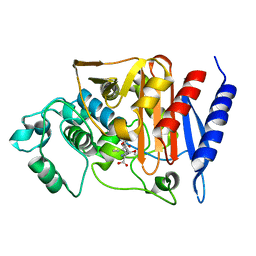 | | 4-Substituted Trinems as Broad Spectrum-Lactamase Inhibitors: Structure-based Design, Synthesis and Biological Activity | | 分子名称: | (1R,4S,7AS)-1-(1-FORMYLPROP-1-EN-1-YL)-4-METHOXY-2,4,5,6,7,7A-HEXAHYDRO-1H-ISOINDOLE-3-CARBOXYLIC ACID, Beta-lactamase | | 著者 | Plantan, I, Selic, L, Mesar, T, Stefanic Anderluh, P, Oblak, M, Prezelj, A, Hesse, L, Andrejasic, M, Vilar, M, Turk, D, Kocijan, A, Prevec, T, Vilfan, G, Kocjan, D, Copar, A, Urleb, U, Solmajer, T. | | 登録日 | 2007-06-13 | | 公開日 | 2007-08-21 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | 4-Substituted Trinems as Broad Spectrum beta-Lactamase Inhibitors: Structure-Based Design, Synthesis, and Biological Activity
J.Med.Chem., 50, 2007
|
|
2Q7Y
 
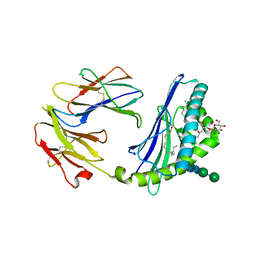 | | Structure of the endogenous iNKT cell ligand iGb3 bound to mCD1d | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | 著者 | Zajonc, D.M, Wilson, I.A, Teyton, L. | | 登録日 | 2007-06-07 | | 公開日 | 2008-04-01 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structures of Mouse CD1d-iGb3 Complex and its Cognate Valpha14 T Cell Receptor Suggest a Model for Dual Recognition of Foreign and Self Glycolipids.
J.Mol.Biol., 377, 2008
|
|
2QU9
 
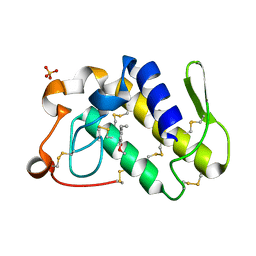 | | Crystal structure of the complex of group II phospholipase A2 with Eugenol | | 分子名称: | 2-methoxy-4-[(1E)-prop-1-en-1-yl]phenol, Phospholipase A2 VRV-PL-VIIIa, SULFATE ION | | 著者 | Kumar, S, Vikram, G, Singh, N, Sinha, M, Sharma, S, Kaur, P, Srinivasan, A, Singh, T.P. | | 登録日 | 2007-08-04 | | 公開日 | 2007-08-14 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Crystal structure of the complex of group II phospholipase A2 with Eugenol
To be Published
|
|
2QV4
 
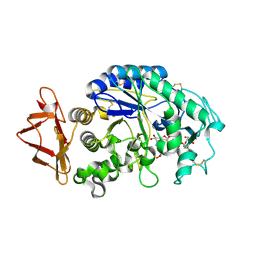 | | Human pancreatic alpha-amylase complexed with nitrite and acarbose | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5R,6S)-4-{[alpha-D-glucopyranosyl-(1->4)-alpha-D-glucopyranosyl-(1->4)-alpha-D-glucopyranosyl]oxy}-5,6-dihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose, CALCIUM ION, ... | | 著者 | Williams, L.K, Maurus, R, Brayer, G.D. | | 登録日 | 2007-08-07 | | 公開日 | 2008-03-25 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.97 Å) | | 主引用文献 | Alternative catalytic anions differentially modulate human alpha-amylase activity and specificity
Biochemistry, 47, 2008
|
|
2VAX
 
 | | Crystal structure of deacetylcephalosporin C acetyltransferase (Cephalosporin C-soak) | | 分子名称: | 4-(3-ACETOXYMETHYL-2-CARBOXY-8-OXO-5-THIA-1-AZA-BICYCLO[4.2.0]OCT-2-EN-7-YLCARBAMOYL)-1-CARBOXY-BUTYL-AMMONIUM, ACETATE ION, ACETYL-COA--DEACETYLCEPHALOSPORIN C ACETYLTRANSFERASE | | 著者 | Lejon, S, Ellis, J, Valegard, K. | | 登録日 | 2007-09-04 | | 公開日 | 2008-09-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The Last Step in Cephalosporin C Formation Revealed: Crystal Structures of Deacetylcephalosporin C Acetyltransferase from Acremonium Chrysogenum in Complexes with Reaction Intermediates.
J.Mol.Biol., 377, 2008
|
|
2VE1
 
 | | Isopenicillin N synthase with substrate analogue AsMCOV (oxygen exposed 1min 20bar) | | 分子名称: | FE (II) ION, ISOPENICILLIN N SYNTHETASE, N^6^-[(1R,2S)-1-({[(1R)-1-carboxy-2-methylpropyl]oxy}carbonyl)-2-sulfanylpropyl]-6-oxo-L-lysine, ... | | 著者 | Ge, W, Clifton, I.J, Adlington, R.M, Baldwin, J.E, Rutledge, P.J. | | 登録日 | 2007-10-15 | | 公開日 | 2008-11-04 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Studies on the Reaction of Isopenicillin N Synthase with a Sterically Demanding Depsipeptide Substrate Analogue.
Chembiochem, 10, 2009
|
|
2W6Z
 
 | |
2W0V
 
 | | Crystal structure of Glmu from Haemophilus influenzae in complex with quinazoline inhibitor 1 | | 分子名称: | 6-(CYCLOPROP-2-EN-1-YLMETHOXY)-2-[6-(CYCLOPROPYLMETHYL)-5-OXO-3,4,5,6-TETRAHYDRO-2,6-NAPHTHYRIDIN-2(1H)-YL]-7-METHOXYQUINAZOLIN-4(3H)-ONE, GLUCOSAMINE-1-PHOSPHATE N-ACETYLTRANSFERASE, SULFATE ION, ... | | 著者 | Mochalkin, I, Melnick, M. | | 登録日 | 2008-10-10 | | 公開日 | 2009-11-17 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.99 Å) | | 主引用文献 | Discovery and Initial Sar of Quinazoline Inhibitors of Glmu from Haemophilus Influenzae
To be Published
|
|
8HW1
 
 | | Far-red light-harvesting complex of Antarctic alga Prasiola crispa | | 分子名称: | (1S)-4-[(1E,3Z,5E,7E,9E,11E,13E,15E,17E)-3-(hydroxymethyl)-7,12,16-trimethyl-18-[(1R,4S)-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]-3,5,5-trimethyl-cyclohex-3-en-1-ol, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, CHLOROPHYLL A, ... | | 著者 | Kosugi, M, Kawasaki, M, Shibata, Y, Moriya, T, Adachi, N, Senda, T. | | 登録日 | 2022-12-28 | | 公開日 | 2023-01-18 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.13 Å) | | 主引用文献 | Uphill energy transfer mechanism for photosynthesis in an Antarctic alga.
Nat Commun, 14, 2023
|
|
2WCM
 
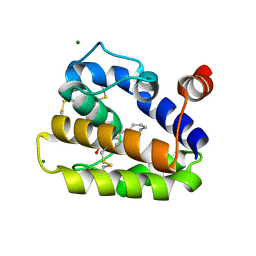 | | Structure of BMori GOBP2 (General Odorant Binding Protein 2) with (10E)-hexadecen-12-yn-1-ol | | 分子名称: | (10E)-hexadec-10-en-12-yn-1-ol, GENERAL ODORANT-BINDING PROTEIN 1, MAGNESIUM ION | | 著者 | Robertson, G, Zhou, J.-J, He, X, Pickett, J.A, Field, L.M, Keep, N.H. | | 登録日 | 2009-03-12 | | 公開日 | 2009-08-11 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Characterisation of Bombyx Mori Odorant-Binding Proteins Reveals that a General Odorant-Binding Protein Discriminates between Sex Pheromone Components.
J.Mol.Biol., 389, 2009
|
|
4U4R
 
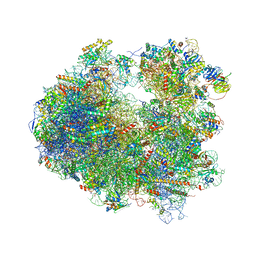 | | Crystal structure of Lactimidomycin bound to the yeast 80S ribosome | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 4-{(2R,5S,6E)-2-hydroxy-5-methyl-7-[(2R,3S,4E,6Z,10E)-3-methyl-12-oxooxacyclododeca-4,6,10-trien-2-yl]-4-oxooct-6-en-1-yl}piperidine-2,6-dione, ... | | 著者 | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | 登録日 | 2014-07-24 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.801 Å) | | 主引用文献 | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
2VAV
 
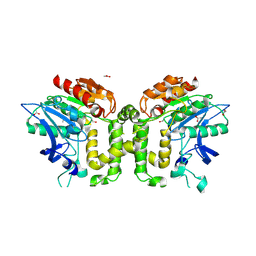 | | Crystal structure of deacetylcephalosporin C acetyltransferase (DAC-Soak) | | 分子名称: | 4-(3-ACETOXYMETHYL-2-CARBOXY-8-OXO-5-THIA-1-AZA-BICYCLO[4.2.0]OCT-2-EN-7-YLCARBAMOYL)-1-CARBOXY-BUTYL-AMMONIUM, ACETATE ION, ACETYL-COA--DEACETYLCEPHALOSPORIN C ACETYLTRANSFERASE | | 著者 | Lejon, S, Ellis, J, Valegard, K. | | 登録日 | 2007-09-04 | | 公開日 | 2008-09-23 | | 最終更新日 | 2018-12-12 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The last step in cephalosporin C formation revealed: crystal structures of deacetylcephalosporin C acetyltransferase from Acremonium chrysogenum in complexes with reaction intermediates.
J. Mol. Biol., 377, 2008
|
|
2V34
 
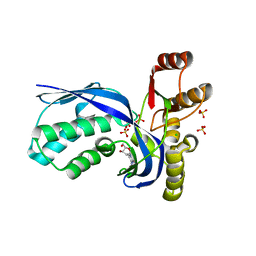 | | IspE in complex with cytidine and ligand | | 分子名称: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, 4-DIPHOSPHOCYTIDYL-2C-METHYL-D-ERYTHRITOL KINASE, CHLORIDE ION, ... | | 著者 | Sgraja, T, Alphey, M.S, Hunter, W.N. | | 登録日 | 2007-06-11 | | 公開日 | 2007-06-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Characterization of Aquifex Aeolicus 4-Diphosphocytidyl-2C-Methyl-D-Erythritol Kinase -Ligand Recognition in a Template for Antimicrobial Drug Discovery.
FEBS J., 275, 2008
|
|
4U53
 
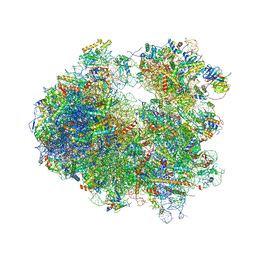 | | Crystal structure of Deoxynivalenol bound to the yeast 80S ribosome | | 分子名称: | (3beta,7alpha)-3,7,15-trihydroxy-12,13-epoxytrichothec-9-en-8-one, 18S ribosomal RNA, 25S ribosomal RNA, ... | | 著者 | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | 登録日 | 2014-07-24 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
2W8Y
 
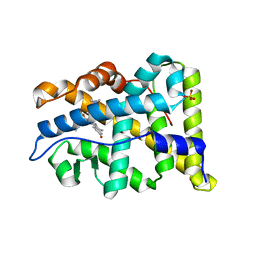 | | RU486 bound to the progesterone receptor in a destabilized agonistic conformation | | 分子名称: | (14beta,17alpha)-17-ethynyl-17-hydroxyestr-4-en-3-one, 1,2-ETHANEDIOL, 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, ... | | 著者 | Raaijmakers, H.C.A, Versteeg, J, Uitdehaag, J.C.M. | | 登録日 | 2009-01-20 | | 公開日 | 2009-04-28 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The X-Ray Structure of Ru486 Bound to the Progesterone Receptor in a Destabilized Agonistic Conformation.
J.Biol.Chem., 284, 2009
|
|
2VBQ
 
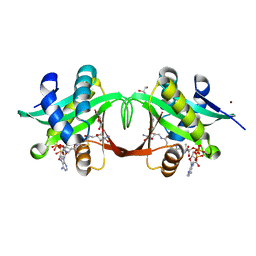 | | Structure of AAC(6')-Iy in complex with bisubstrate analog CoA-S- monomethyl-acetylneamine. | | 分子名称: | (3R,9Z)-17-[(2R,3S,4R,5R,6R)-5-amino-6-{[(1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl]oxy}-3,4-dihydroxytetrahydro-2H-pyran-2-yl]-3-hydroxy-2,2-dimethyl-4,8,15-trioxo-12-thia-5,9,16-triazaheptadec-9-en-1-yl [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, AMINOGLYCOSIDE 6'-N-ACETYLTRANSFERASE, GLYCEROL, ... | | 著者 | Vetting, M.W, Magalhaes, M.L, Freiburger, L, Gao, F, Auclair, K, Blanchard, J.S. | | 登録日 | 2007-09-14 | | 公開日 | 2008-01-08 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Kinetic and Structural Analysis of Bisubstrate Inhibition of the Salmonella Enterica Aminoglycoside 6'-N-Acetyltransferase.
Biochemistry, 47, 2008
|
|
2W4Q
 
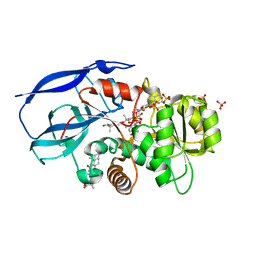 | | Crystal structure of human zinc-binding alcohol dehydrogenase 1 (ZADH1) in ternary complex with NADP and 18beta-glycyrrhetinic acid | | 分子名称: | (3BETA,5BETA,14BETA)-3-HYDROXY-11-OXOOLEAN-12-EN-29-OIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROSTAGLANDIN REDUCTASE 2, ... | | 著者 | Shafqat, N, Yue, W.W, Ugochukwu, E, Picaud, S, Niesen, F, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | 登録日 | 2008-11-30 | | 公開日 | 2009-01-20 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of Human Zinc-Binding Alcohol Dehydrogenase 1
To be Published
|
|
2WTX
 
 | | Insight into the mechanism of enzymatic glycosyltransfer with retention through the synthesis and analysis of bisubstrate glycomimetics of trehalose-6-phosphate synthase | | 分子名称: | 1,2-ETHANEDIOL, ALPHA, ALPHA-TREHALOSE-PHOSPHATE SYNTHASE [UDP-FORMING], ... | | 著者 | Errey, J.C, Lee, S.S, Gibson, R.P, Martinez-Fleites, C, Barry, C.S, Jung, P.M.J, OSullivan, A, Davis, B.G, Davies, G.J. | | 登録日 | 2009-09-25 | | 公開日 | 2010-02-23 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mechanistic Insight Into Enzymatic Glycosyl Transfer with Retention of Configuration Through Analysis of Glycomimetic Inhibitors.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2VAO
 
 | |
4Q27
 
 | |
7LI5
 
 | | Crystal Structure Analysis of human TEAD1 | | 分子名称: | 1-[(3R,4R)-3-[4-(pyridin-3-yl)-1H-1,2,3-triazol-1-yl]-4-{[4-(trifluoromethyl)phenyl]methoxy}pyrrolidin-1-yl]prop-2-en-1-one, SULFATE ION, Transcriptional enhancer factor TEF-1 | | 著者 | Seo, H.-S, Dhe-Paganon, S. | | 登録日 | 2021-01-26 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Crystal Structure Analysis of human TEAD1
To Be Published
|
|
