6YN0
 
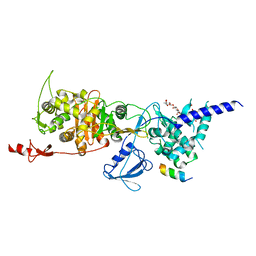 | | Structure of E. coli PBP1b with a FtsN peptide activating transglycosylase activity | | 分子名称: | Cell division protein FtsN, MOENOMYCIN, Penicillin-binding protein 1B | | 著者 | Kerff, F, Terrak, M, Boes, A, Herman, H, Charlier, P. | | 登録日 | 2020-04-10 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The bacterial cell division protein fragment E FtsN binds to and activates the major peptidoglycan synthase PBP1b.
J.Biol.Chem., 295, 2020
|
|
6TIX
 
 | |
6TII
 
 | |
7ASS
 
 | |
7KIS
 
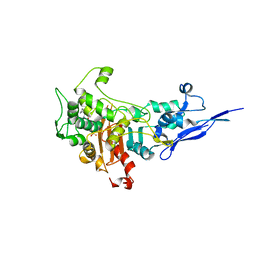 | | Crystal structure of Pseudomonas aeruginosa PBP2 in complex with WCK 5153 | | 分子名称: | (2S,5R)-1-formyl-N'-[(3R)-pyrrolidine-3-carbonyl]-5-[(sulfooxy)amino]piperidine-2-carbohydrazide, CHLORIDE ION, Peptidoglycan D,D-transpeptidase MrdA | | 著者 | Rajavel, M, van den Akker, F. | | 登録日 | 2020-10-24 | | 公開日 | 2021-01-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.869 Å) | | 主引用文献 | Structural Characterization of Diazabicyclooctane beta-Lactam "Enhancers" in Complex with Penicillin-Binding Proteins PBP2 and PBP3 of Pseudomonas aeruginosa.
Mbio, 12, 2021
|
|
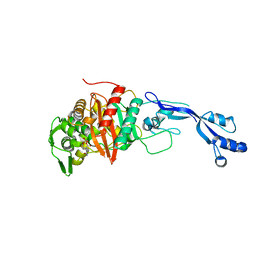
7KIW
 
 | |
7KIV
 
 | |
7KIT
 
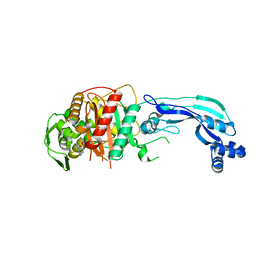 | |
6TUD
 
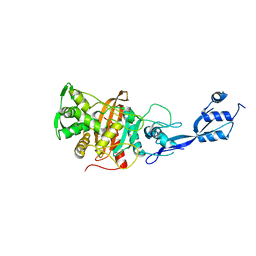 | |
6XV5
 
 | |
7KHY
 
 | | Crystal structure of OXA-163 K73A in complex with meropenem | | 分子名称: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-lactamase, ... | | 著者 | Palzkill, T, Hu, L, Sankaran, B, Prasad, B.V.V. | | 登録日 | 2020-10-22 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Mechanistic Basis of OXA-48-like beta-Lactamases' Hydrolysis of Carbapenems.
Acs Infect Dis., 7, 2021
|
|
7KHQ
 
 | | Crystal structure of OXA-48 K73A in complex with meropenem | | 分子名称: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase, CHLORIDE ION, ... | | 著者 | Palzkill, T, Hu, L, Sankaran, B, Prasad, B.V.V. | | 登録日 | 2020-10-21 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mechanistic Basis of OXA-48-like beta-Lactamases' Hydrolysis of Carbapenems.
Acs Infect Dis., 7, 2021
|
|
7KH9
 
 | | Crystal structure of OXA-48 K73A in complex with imipenem | | 分子名称: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Beta-lactamase, CHLORIDE ION | | 著者 | Palzkill, T, Hu, L, Sankaran, B, Prasad, B.V.V. | | 登録日 | 2020-10-20 | | 公開日 | 2021-02-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Mechanistic Basis of OXA-48-like beta-Lactamases' Hydrolysis of Carbapenems.
Acs Infect Dis., 7, 2021
|
|
7KHZ
 
 | | Crystal structure of OXA-163 K73A in complex with imipenem | | 分子名称: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, Beta-lactamase, CHLORIDE ION, ... | | 著者 | Palzkill, T, Hu, L, Sankaran, B, Prasad, B.V.V. | | 登録日 | 2020-10-22 | | 公開日 | 2021-02-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Mechanistic Basis of OXA-48-like beta-Lactamases' Hydrolysis of Carbapenems.
Acs Infect Dis., 7, 2021
|
|
7LC4
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with gamma-lactam YU253911 | | 分子名称: | 1-[(2S)-2-{[(2Z)-2-(2-amino-5-chloro-1,3-thiazol-4-yl)-2-{[(2-carboxypropan-2-yl)oxy]imino}acetyl]amino}-3-oxopropyl]-4-{[2-(5,6-dihydroxy-1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl]carbamoyl}-2,5-dihydro-1H-pyrazole-3-carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | 著者 | van den Akker, F, Kumar, V. | | 登録日 | 2021-01-09 | | 公開日 | 2021-04-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | A gamma-lactam siderophore antibiotic effective against multidrug-resistant Pseudomonas aeruginosa, Klebsiella pneumoniae, and Acinetobacter spp.
Eur.J.Med.Chem., 220, 2021
|
|
7LQ6
 
 | | CryoEM structure of Escherichia coli PBP1b | | 分子名称: | Penicillin-binding protein 1B | | 著者 | Caveney, N.A, Workman, S.D, Yan, R, Atkinson, C.E, Yu, Z, Strynadka, N.C.J. | | 登録日 | 2021-02-13 | | 公開日 | 2021-05-26 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.28 Å) | | 主引用文献 | CryoEM structure of the antibacterial target PBP1b at 3.3 angstrom resolution.
Nat Commun, 12, 2021
|
|
7JWL
 
 | | Crystal Structure of Pseudomonas aeruginosa Penicillin Binding Protein 3 (PAE-PBP3) bound to ETX0462 | | 分子名称: | CHLORIDE ION, ETX0462 (Bound form), Peptidoglycan D,D-transpeptidase FtsI | | 著者 | Mayclin, S.J, Abendroth, J, Horanyi, P.S, Sylvester, M, Wu, X, Shapiro, A, Moussa, S, Durand-Reville, T.F. | | 登録日 | 2020-08-25 | | 公開日 | 2021-05-26 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Rational design of a new antibiotic class for drug-resistant infections.
Nature, 597, 2021
|
|
7AUX
 
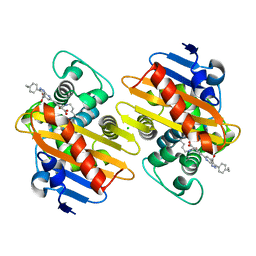 | | Crystal structure of OXA-48 beta-lactamase in the complex with the inhbitor ID2 | | 分子名称: | 6-(4-carboxyphenyl)-3-(4-ethylphenyl)-2~{H}-pyrazolo[3,4-b]pyridine-4-carboxylic acid, Beta-lactamase, CHLORIDE ION | | 著者 | Pochetti, G, Montanari, R, Capelli, D, Garofalo, B, Ombrato, R. | | 登録日 | 2020-11-03 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Discovery of Novel Chemical Series of OXA-48 beta-Lactamase Inhibitors by High-Throughput Screening.
Pharmaceuticals, 14, 2021
|
|
7AW5
 
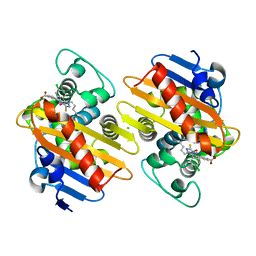 | | Crystal structure of OXA-48 beta-lactamase in the complex with the inhibitor ID3 | | 分子名称: | 4-[(~{E})-[3-(4-chlorophenyl)-5-sulfanylidene-1~{H}-1,2,4-triazol-4-yl]iminomethyl]benzoic acid, Beta-lactamase, CHLORIDE ION | | 著者 | Pochetti, G, Montanari, R, Capelli, D, Garofalo, B, Ombrato, R. | | 登録日 | 2020-11-06 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Discovery of Novel Chemical Series of OXA-48 beta-Lactamase Inhibitors by High-Throughput Screening.
Pharmaceuticals, 14, 2021
|
|
6XQV
 
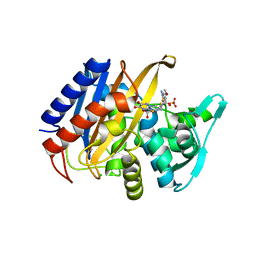 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae in a pre-acylation complex with ceftriaxone | | 分子名称: | CHLORIDE ION, Ceftriaxone, Probable peptidoglycan D,D-transpeptidase PenA, ... | | 著者 | Fenton, B.A, Zhou, P, Davies, C. | | 登録日 | 2020-07-10 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6XQZ
 
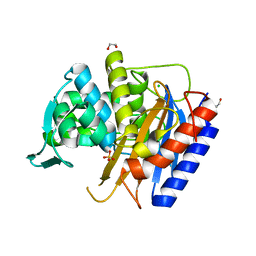 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae at pH 7.5 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptidoglycan D,D-transpeptidase PenA, ... | | 著者 | Fenton, B.A, Zhou, P, Davies, C. | | 登録日 | 2020-07-10 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.04 Å) | | 主引用文献 | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6XQY
 
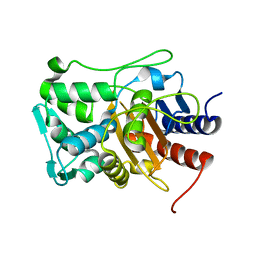 | |
6XQX
 
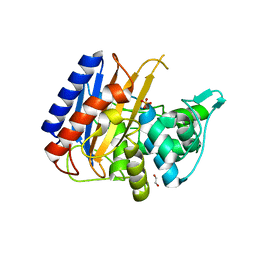 | | Crystal structure of the catalytic domain of PBP2 S310A from Neisseria gonorrhoeae with the H514A mutation at pH 7.5 | | 分子名称: | 1,2-ETHANEDIOL, Probable peptidoglycan D,D-transpeptidase PenA, SULFATE ION | | 著者 | Fenton, B.A, Zhou, P, Davies, C. | | 登録日 | 2020-07-10 | | 公開日 | 2021-07-21 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Mutations in PBP2 from ceftriaxone-resistant Neisseria gonorrhoeae alter the dynamics of the beta 3-beta 4 loop to favor a low-affinity drug-binding state.
J.Biol.Chem., 297, 2021
|
|
6ZRG
 
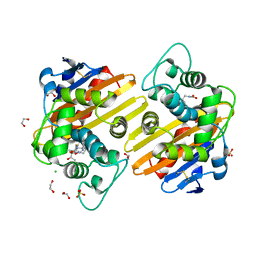 | | Crystal structure of OXA-10loop48 in complex with hydrolyzed doripenem | | 分子名称: | (4R,5S)-3-{[(3S,5S)-5-({[amino(dihydroxy)methyl]amino}methyl)pyrrolidin-3-yl]sulfanyl}-5-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | 著者 | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Docquier, J.D, Mangani, S. | | 登録日 | 2020-07-13 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.74 Å) | | 主引用文献 | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6ZRJ
 
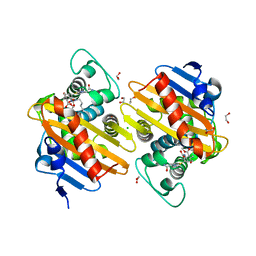 | | Crystal structure of class D Beta-lactamase OXA-48 in complex with ertapenem | | 分子名称: | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | 著者 | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Docquier, J.D, Mangani, S. | | 登録日 | 2020-07-13 | | 公開日 | 2021-07-21 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
