5IVF
 
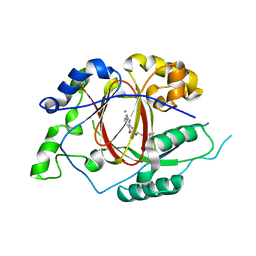 | | Linked KDM5A Jmj Domain Bound to the Inhibitor N10 8-(1-methyl-1H-imidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol | | 分子名称: | 8-(1-methyl-1H-imidazol-4-yl)-2-(4,4,4-trifluorobutoxy)pyrido[3,4-d]pyrimidin-4-ol, Lysine-specific demethylase 5A, MANGANESE (II) ION | | 著者 | Horton, J.R, Cheng, X. | | 登録日 | 2016-03-20 | | 公開日 | 2016-07-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.683 Å) | | 主引用文献 | Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds.
Cell Chem Biol, 23, 2016
|
|
5XS8
 
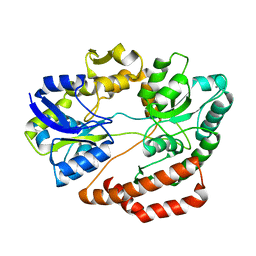 | | Crystal structure of solute-binding protein complexed with unsaturated chondroitin disaccharide with two sulfate groups at C-4 and C-6 positions of GalNAc | | 分子名称: | 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4,6-di-O-sulfo-beta-D-galactopyranose, CALCIUM ION, Extracellular solute-binding protein family 1 | | 著者 | Oiki, S, Kamochi, R, Mikami, B, Murata, K, Hashimoto, W. | | 登録日 | 2017-06-12 | | 公開日 | 2018-01-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.952 Å) | | 主引用文献 | Alternative substrate-bound conformation of bacterial solute-binding protein involved in the import of mammalian host glycosaminoglycans.
Sci Rep, 7, 2017
|
|
1BS5
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM | | 分子名称: | PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ZINC ION | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BSS
 
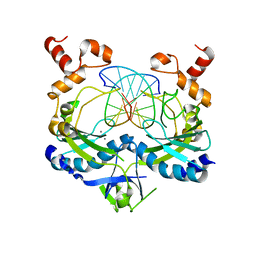 | | ECORV-T93A/DNA/CA2+ | | 分子名称: | 5'-D(*AP*AP*AP*GP*AP*TP*AP*TP*CP*TP*T)-3', CALCIUM ION, ECORV ENDONUCLEASE | | 著者 | Perona, J.J, Horton, N.C. | | 登録日 | 1998-08-30 | | 公開日 | 1998-09-02 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Metal ion-mediated substrate-assisted catalysis in type II restriction endonucleases
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
7A0K
 
 | |
1BTX
 
 | |
1BUA
 
 | |
7AC7
 
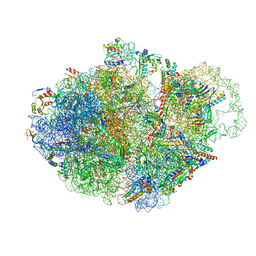 | | Structure of accomodated trans-translation complex on E. Coli stalled ribosome. | | 分子名称: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | 著者 | Guyomar, C, D'Urso, G, Chat, S, Giudice, E, Gillet, R. | | 登録日 | 2020-09-10 | | 公開日 | 2021-08-18 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (3.08 Å) | | 主引用文献 | Structures of tmRNA and SmpB as they transit through the ribosome.
Nat Commun, 12, 2021
|
|
1BV4
 
 | | APO-MANNOSE-BINDING PROTEIN-C | | 分子名称: | PROTEIN (MANNOSE-BINDING PROTEIN-C) | | 著者 | Ng, K.K.-S, Weis, W.I. | | 登録日 | 1998-09-22 | | 公開日 | 1999-01-13 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Ca2+-dependent structural changes in C-type mannose-binding proteins.
Biochemistry, 37, 1998
|
|
1BVO
 
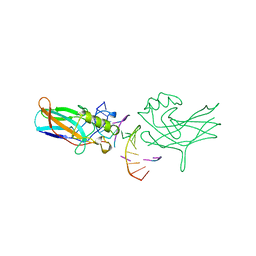 | | DORSAL HOMOLOGUE GAMBIF1 BOUND TO DNA | | 分子名称: | DNA DUPLEX, TRANSCRIPTION FACTOR GAMBIF1 | | 著者 | Cramer, P, Varrot, A, Barillas-Mury, C, Kafatos, F.C, Mueller, C.W. | | 登録日 | 1998-09-16 | | 公開日 | 1999-07-12 | | 最終更新日 | 2017-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structure of the specificity domain of the Dorsal homologue Gambif1 bound to DNA.
Structure Fold.Des., 7, 1999
|
|
1BVV
 
 | |
7AE5
 
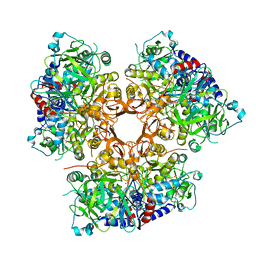 | |
5EUJ
 
 | | PYRUVATE DECARBOXYLASE FROM ZYMOBACTER PALMAE | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, Pyruvate decarboxylase, ... | | 著者 | Buddrus, L, Crennell, S.J, Leak, D.J, Danson, M.J, Andrews, E.S.V, Arcus, V.L. | | 登録日 | 2015-11-18 | | 公開日 | 2016-09-14 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structure of pyruvate decarboxylase from Zymobacter palmae.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
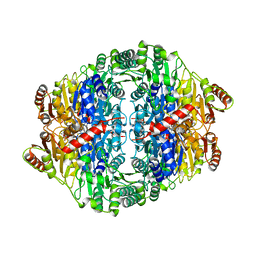
5F38
 
 | | X-ray crystal structure of a thiolase from Escherichia coli at 1.8 A resolution | | 分子名称: | 1,2-ETHANEDIOL, Acetyl-CoA acetyltransferase, COENZYME A, ... | | 著者 | Ithayaraja, M, Neelanjana, J, Wierenga, R, Savithri, H.S, Murthy, M.R.N. | | 登録日 | 2015-12-02 | | 公開日 | 2016-07-13 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of a thiolase from Escherichia coli at 1.8 angstrom resolution.
Acta Crystallogr.,Sect.F, 72, 2016
|
|
1BWB
 
 | | HIV-1 PROTEASE (V82F/I84V) DOUBLE MUTANT COMPLEXED WITH SD146 OF DUPONT PHARMACEUTICALS | | 分子名称: | PROTEIN (HIV-1 PROTEASE), [4R-(4ALPHA,5ALPHA,6ALPHA,7ALPHA)]-3,3'-{{TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-DIYL]BIS(METHYLENE)]BIS[N-1H-BENZIMIDAZOL-2-YLBENZAMIDE] | | 著者 | Ala, P, Chang, C.H. | | 登録日 | 1998-09-22 | | 公開日 | 1998-09-30 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities.
Biochemistry, 37, 1998
|
|
1BS8
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | 分子名称: | PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | 著者 | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | 登録日 | 1998-09-01 | | 公開日 | 1999-08-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BSO
 
 | | 12-BROMODODECANOIC ACID BINDS INSIDE THE CALYX OF BOVINE BETA-LACTOGLOBULIN | | 分子名称: | 12-BROMODODECANOIC ACID, PROTEIN (BOVINE BETA-LACTOGLOBULIN A) | | 著者 | Qin, B.Y, Creamer, L.K, Baker, E.N, Jameson, G.B. | | 登録日 | 1998-08-29 | | 公開日 | 1999-09-02 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | 12-Bromododecanoic acid binds inside the calyx of bovine beta-lactoglobulin.
FEBS Lett., 438, 1998
|
|
1BWI
 
 | | THE 1.8 A STRUCTURE OF MICROBATCH OIL DROP GROWN TETRAGONAL HEN EGG WHITE LYSOZYME | | 分子名称: | PROTEIN (LYSOZYME) | | 著者 | Dong, J, Boggon, T.J, Chayen, N.E, Raftery, J, Bi, R.C. | | 登録日 | 1998-09-24 | | 公開日 | 1998-09-30 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Bound-solvent structures for microgravity-, ground control-, gel- and microbatch-grown hen egg-white lysozyme crystals at 1.8 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BWK
 
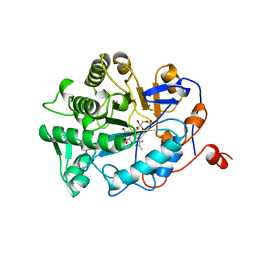 | | OLD YELLOW ENZYME (OYE1) MUTANT H191N | | 分子名称: | FLAVIN MONONUCLEOTIDE, PROTEIN (NADPH DEHYDROGENASE 1) | | 著者 | Brown, B.J, Deng, Z, Karplus, P.A, Massey, V. | | 登録日 | 1998-09-24 | | 公開日 | 1998-09-30 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | On the active site of Old Yellow Enzyme. Role of histidine 191 and asparagine 194.
J.Biol.Chem., 273, 1998
|
|
1BTJ
 
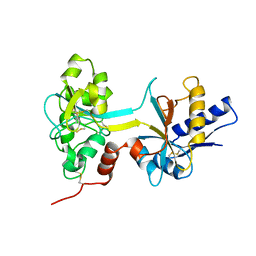 | | HUMAN SERUM TRANSFERRIN, RECOMBINANT N-TERMINAL LOBE, APO FORM, CRYSTAL FORM 2 | | 分子名称: | PROTEIN (SERUM TRANSFERRIN) | | 著者 | Jeffrey, P.D, Bewley, M.C, Macgillivray, R.T.A, Mason, A.B, Woodworth, R.C, Baker, E.N. | | 登録日 | 1998-09-01 | | 公開日 | 1999-01-13 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Ligand-induced conformational change in transferrins: crystal structure of the open form of the N-terminal half-molecule of human transferrin.
Biochemistry, 37, 1998
|
|
1BWZ
 
 | |
5EZM
 
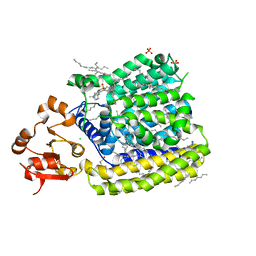 | | Crystal Structure of ArnT from Cupriavidus metallidurans in the apo state | | 分子名称: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferases of PMT family, CHLORIDE ION, ... | | 著者 | Petrou, V.I, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2015-11-26 | | 公開日 | 2016-02-17 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Structures of aminoarabinose transferase ArnT suggest a molecular basis for lipid A glycosylation.
Science, 351, 2016
|
|
1BU8
 
 | | RAT PANCREATIC LIPASE RELATED PROTEIN 2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (PANCREATIC LIPASE RELATED PROTEIN 2) | | 著者 | Roussel, A, Cambillau, C. | | 登録日 | 1998-09-14 | | 公開日 | 1998-12-23 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure and activity of rat pancreatic lipase-related protein 2.
J.Biol.Chem., 273, 1998
|
|
1BXT
 
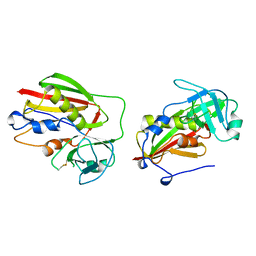 | |
1BUL
 
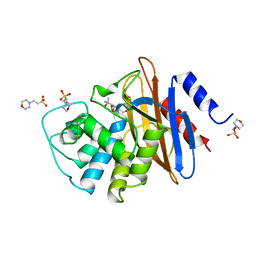 | | 6ALPHA-(HYDROXYPROPYL)PENICILLANATE ACYLATED ON NMC-A BETA-LACTAMASE FROM ENTEROBACTER CLOACAE | | 分子名称: | 2-(1-CARBOXY-2-HYDROXY-2-METHYL-PROPYL)-5,5-DIMETHYL-THIAZOLIDINE-4-CARBOXYLIC ACID, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, NMC-A BETA-LACTAMASE | | 著者 | Mourey, L, Swaren, P, Miyashita, K, Bulychev, A, Mobashery, S, Samama, J.P. | | 登録日 | 1998-09-04 | | 公開日 | 1998-12-30 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Inhibition of the Nmc-A B-Lactamase by a Penicillanic Acid Derivative, and the Structural Bases for the Increase in Substrate Profile of This Antibiotic Resistance Enzyme
J.Am.Chem.Soc., 120, 1998
|
|
