9F99
 
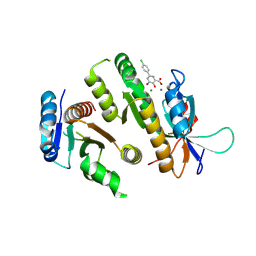 | | Crystal structure of MUS81-EME1 bound by compound 10. | | 分子名称: | 2-(4-chlorophenyl)-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | 著者 | Collie, G.W. | | 登録日 | 2024-05-07 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.803 Å) | | 主引用文献 | Fragment-Based Discovery of Novel MUS81 Inhibitors
Acs Med.Chem.Lett., 2024
|
|
9F98
 
 | | Crystal structure of MUS81-EME1, apo form. | | 分子名称: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81 | | 著者 | Collie, G.W. | | 登録日 | 2024-05-07 | | 公開日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Fragment-Based Discovery of Novel MUS81 Inhibitors
Acs Med.Chem.Lett., 2024
|
|
9F9L
 
 | | Crystal structure of MUS81-EME1 bound by compound 16. | | 分子名称: | 2-[2-[4-(cyanomethyl)phenyl]phenyl]-5-oxidanyl-6-oxidanylidene-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | 著者 | Collie, G.W. | | 登録日 | 2024-05-07 | | 公開日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Fragment-Based Discovery of Novel MUS81 Inhibitors
Acs Med.Chem.Lett., 2024
|
|
9F9K
 
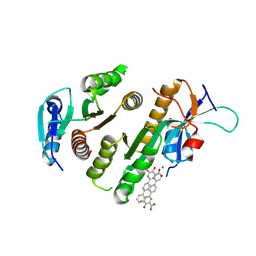 | | Crystal structure of MUS81-EME1 bound by compound 15. | | 分子名称: | 5-oxidanyl-6-oxidanylidene-2-(4-phenylphenyl)-1H-pyrimidine-4-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | 著者 | Collie, G.W. | | 登録日 | 2024-05-07 | | 公開日 | 2024-07-03 | | 実験手法 | X-RAY DIFFRACTION (2.728 Å) | | 主引用文献 | Fragment-Based Discovery of Novel MUS81 Inhibitors
Acs Med.Chem.Lett., 2024
|
|
5XJC
 
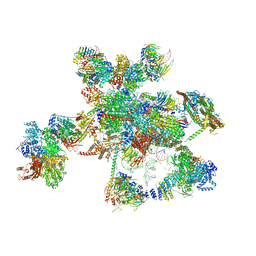 | | Cryo-EM structure of the human spliceosome just prior to exon ligation at 3.6 angstrom | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Zhang, X, Yan, C, Hang, J, Finci, I.L, Lei, J, Shi, Y. | | 登録日 | 2017-04-30 | | 公開日 | 2017-07-05 | | 最終更新日 | 2020-10-14 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | An Atomic Structure of the Human Spliceosome
Cell, 169, 2017
|
|
9F9M
 
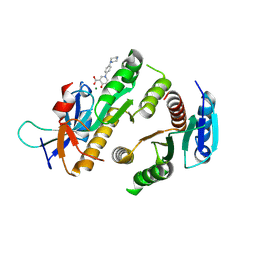 | | Crystal structure of MUS81-EME1 bound by compound 21. | | 分子名称: | 5-oxidanyl-4-oxidanylidene-1-(4-piperazin-1-ylphenyl)pyridine-3-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | 著者 | Collie, G.W. | | 登録日 | 2024-05-08 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (2.469 Å) | | 主引用文献 | Fragment-Based Discovery of Novel MUS81 Inhibitors
Acs Med.Chem.Lett., 2024
|
|
6BFI
 
 | |
4H1E
 
 | |
5Y04
 
 | |
1PGS
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF PNGASE F, A GLYCOSYLASPARAGINASE FROM FLAVOBACTERIUM MENINGOSEPTICUM | | 分子名称: | PEPTIDE-N(4)-(N-ACETYL-BETA-D-GLUCOSAMINYL)ASPARAGINE AMIDASE F | | 著者 | Norris, G.E, Stillman, T.J, Anderson, B.F, Baker, E.N. | | 登録日 | 1994-10-06 | | 公開日 | 1995-01-26 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The three-dimensional structure of PNGase F, a glycosylasparaginase from Flavobacterium meningosepticum.
Structure, 2, 1994
|
|
1PNF
 
 | | PNGASE F COMPLEX WITH DI-N-ACETYLCHITOBIOSE | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, PEPTIDE-N(4)-(N-ACETYL-BETA-D-GLUCOSAMINYL)ASPARAGINE AMIDASE F, SULFATE ION | | 著者 | Van Roey, P, Kuhn, P. | | 登録日 | 1995-10-11 | | 公開日 | 1996-03-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Active site and oligosaccharide recognition residues of peptide-N4-(N-acetyl-beta-D-glucosaminyl)asparagine amidase F.
J.Biol.Chem., 270, 1995
|
|
1PPE
 
 | |
1I7X
 
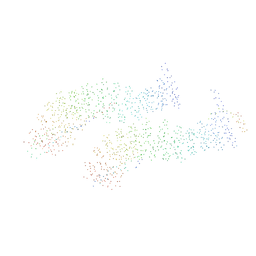 | | BETA-CATENIN/E-CADHERIN COMPLEX | | 分子名称: | BETA-CATENIN, EPITHELIAL-CADHERIN | | 著者 | Huber, A.H, Weis, W.I. | | 登録日 | 2001-03-10 | | 公開日 | 2001-05-16 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin.
Cell(Cambridge,Mass.), 105, 2001
|
|
7UKK
 
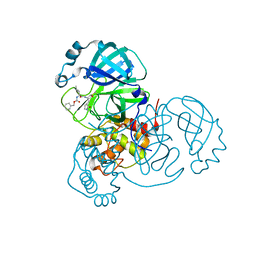 | |
1U2A
 
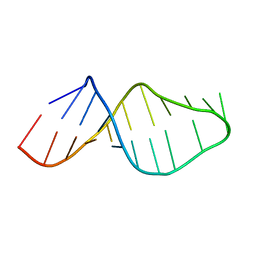 | |
1LM7
 
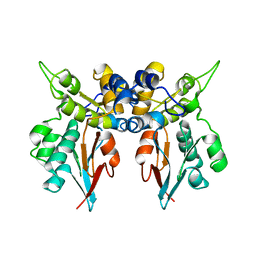 | | Structures of two intermediate filament-binding fragments of desmoplakin reveal a unique repeat motif structure | | 分子名称: | subdomain of Desmoplakin Carboxy-Terminal domain (DPCT) | | 著者 | Choi, H.J, Park-Snyder, S, Pascoe, L.T, Green, K.J, Weis, W.I. | | 登録日 | 2002-04-30 | | 公開日 | 2002-07-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structures of two intermediate filament-binding fragments of desmoplakin reveal a unique repeat motif structure.
Nat.Struct.Biol., 9, 2002
|
|
1PAX
 
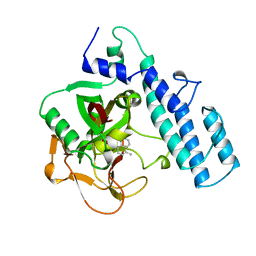 | |
5GNI
 
 | |
1I7W
 
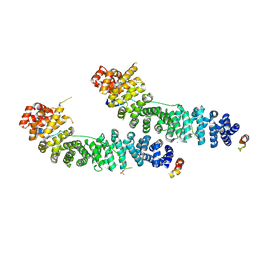 | | BETA-CATENIN/PHOSPHORYLATED E-CADHERIN COMPLEX | | 分子名称: | BETA-CATENIN, CHLORIDE ION, EPITHELIAL-CADHERIN, ... | | 著者 | Huber, A.H, Weis, W.I. | | 登録日 | 2001-03-10 | | 公開日 | 2001-05-09 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The structure of the beta-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by beta-catenin.
Cell(Cambridge,Mass.), 105, 2001
|
|
3RF2
 
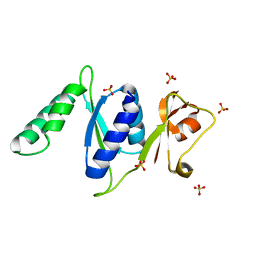 | |
5FD1
 
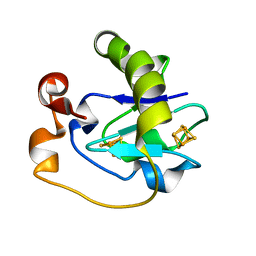 | |
1P4E
 
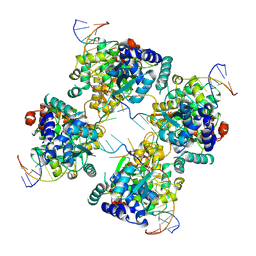 | | Flpe W330F mutant-DNA Holliday Junction Complex | | 分子名称: | 33-MER, 5'-D(*TP*AP*AP*GP*TP*TP*CP*CP*TP*AP*TP*TP*C)-3', 5'-D(*TP*TP*TP*AP*AP*AP*AP*GP*AP*AP*TP*AP*GP*GP*AP*AP*CP*TP*TP*C)-3', ... | | 著者 | Rice, P.A, Chen, Y. | | 登録日 | 2003-04-23 | | 公開日 | 2003-05-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | The role of the conserved Trp330 in Flp-mediated recombination. Functional and structural analysis
J.Biol.Chem., 278, 2003
|
|
3ZBO
 
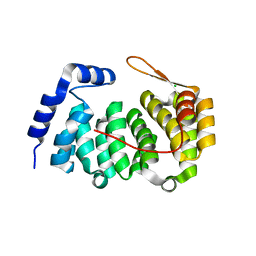 | | A new family of proteins related to the HEAT-like repeat DNA glycosylases with affinity for branched DNA structures | | 分子名称: | ALKF, CHLORIDE ION | | 著者 | Backe, P.H, Simm, R, Laerdahl, J.K, Dalhus, B, Fagerlund, A, Okstad, O.A, Rognes, T, Alseth, I, Kolsto, A.-B, Bjoras, M. | | 登録日 | 2012-11-12 | | 公開日 | 2013-05-15 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | A New Family of Proteins Related to the Heat-Like Repeat DNA Glycosylases with Affinity for Branched DNA Structures.
J.Struct.Biol., 183, 2013
|
|
1LM5
 
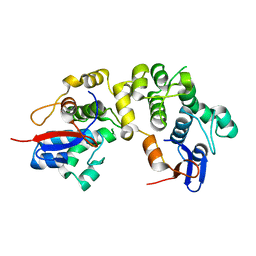 | | Structures of two intermediate filament-binding fragments of desmoplakin reveal a unique repeat motif structure | | 分子名称: | subdomain of Desmoplakin Carboxy-Terminal domain (DPCT) | | 著者 | Choi, H.J, Park-Snyder, S, Pascoe, L.T, Green, K.J, Weis, W.I. | | 登録日 | 2002-04-30 | | 公開日 | 2002-07-31 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structures of two intermediate filament-binding fragments of desmoplakin reveal a unique repeat motif structure.
Nat.Struct.Biol., 9, 2002
|
|
4BOO
 
 | |
