4MFD
 
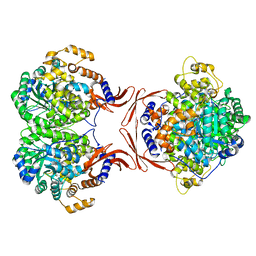 | |
4MFE
 
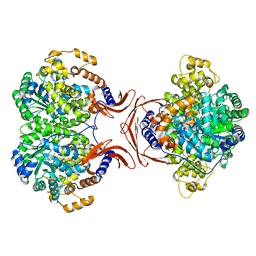 | |
4MIM
 
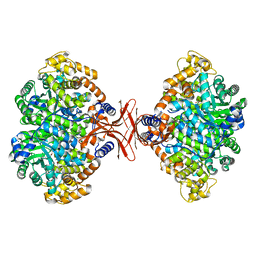 | |
2B3I
 
 | | NMR SOLUTION STRUCTURE OF PLASTOCYANIN FROM THE PHOTOSYNTHETIC PROKARYOTE, PROCHLOROTHRIX HOLLANDICA (19 STRUCTURES) | | 分子名称: | COPPER (I) ION, PROTEIN (PLASTOCYANIN) | | 著者 | Babu, C.R, Volkman, B.F, Bullerjahn, G.S. | | 登録日 | 1998-12-11 | | 公開日 | 1999-04-27 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of plastocyanin from the photosynthetic prokaryote, Prochlorothrix hollandica.
Biochemistry, 38, 1999
|
|
9E8K
 
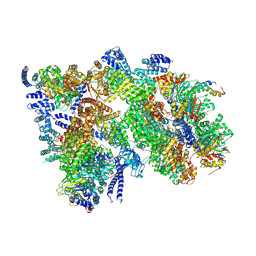 | |
7QO6
 
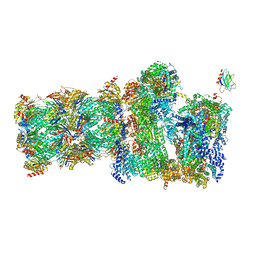 | | 26S proteasome Rpt1-RK -Ubp6-UbVS complex in the s2 state | | 分子名称: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | 著者 | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, T.C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | 登録日 | 2021-12-23 | | 公開日 | 2022-03-16 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (6.3 Å) | | 主引用文献 | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
7QO5
 
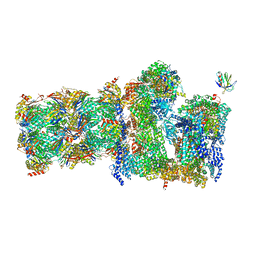 | | 26S proteasome Rpt1-RK -Ubp6-UbVS complex in the si state | | 分子名称: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit 4 homolog, 26S proteasome regulatory subunit 6A, ... | | 著者 | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, T.C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | 登録日 | 2021-12-23 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-03-15 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
7QO3
 
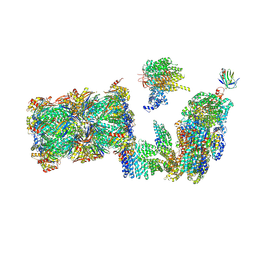 | | Structure of the 26S proteasome-Ubp6 complex in the si state (Core Particle and Lid) | | 分子名称: | 26S proteasome complex subunit SEM1, 26S proteasome regulatory subunit RPN1, 26S proteasome regulatory subunit RPN10, ... | | 著者 | Hung, K.Y.S, Klumpe, S, Eisele, M.R, Elsasser, S, Geng, T.T, Cheng, T.C, Joshi, T, Rudack, T, Sakata, E, Finley, D. | | 登録日 | 2021-12-23 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (6.1 Å) | | 主引用文献 | Allosteric control of Ubp6 and the proteasome via a bidirectional switch.
Nat Commun, 13, 2022
|
|
5XSW
 
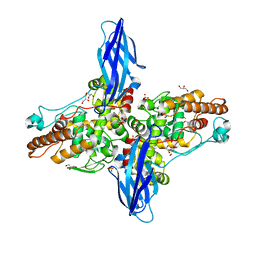 | |
6WJN
 
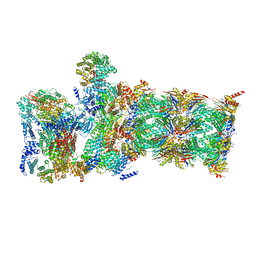 | |
5YF4
 
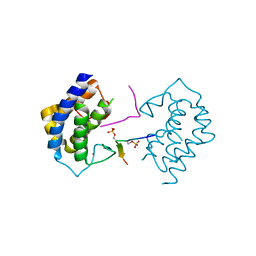 | | A kinase complex MST4-MOB4 | | 分子名称: | MOB-like protein phocein, Peptide from Serine/threonine-protein kinase 26, ZINC ION | | 著者 | Chen, M, Zhou, Z.C. | | 登録日 | 2017-09-20 | | 公開日 | 2018-08-15 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.897 Å) | | 主引用文献 | The MST4-MOB4 complex disrupts the MST1-MOB1 complex in the Hippo-YAP pathway and plays a pro-oncogenic role in pancreatic cancer.
J. Biol. Chem., 293, 2018
|
|
2AG9
 
 | | Crystal Structure of the Y137S mutant of GM2-Activator Protein | | 分子名称: | Ganglioside GM2 activator, ISOPROPYL ALCOHOL, MYRISTIC ACID | | 著者 | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | 登録日 | 2005-07-26 | | 公開日 | 2005-10-25 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
8GK8
 
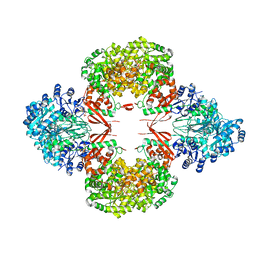 | | R21A Staphylococcus aureus pyruvate carboxylase | | 分子名称: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ACETYL COENZYME *A, COENZYME A, ... | | 著者 | Laseke, A.J, St.Maurice, M. | | 登録日 | 2023-03-17 | | 公開日 | 2023-08-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.68 Å) | | 主引用文献 | Allosteric Site at the Biotin Carboxylase Dimer Interface Mediates Activation and Inhibition in Staphylococcus aureus Pyruvate Carboxylase.
Biochemistry, 62, 2023
|
|
3BG5
 
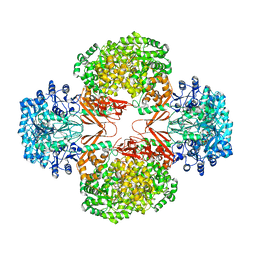 | | Crystal Structure of Staphylococcus Aureus Pyruvate Carboxylase | | 分子名称: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | 著者 | Xiang, S, Tong, L. | | 登録日 | 2007-11-26 | | 公開日 | 2008-02-26 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal structures of human and Staphylococcus aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction.
Nat.Struct.Mol.Biol., 15, 2008
|
|
6WJD
 
 | |
2AG4
 
 | | Crystal Structure Analysis of GM2-activator protein complexed with phosphatidylcholine | | 分子名称: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, Ganglioside GM2 activator, ISOPROPYL ALCOHOL, ... | | 著者 | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | 登録日 | 2005-07-26 | | 公開日 | 2005-10-25 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
2AG2
 
 | | Crystal Structure Analysis of GM2-activator protein complexed with Phosphatidylcholine | | 分子名称: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 2-(((R)-2,3-DIHYDROXYPROPYL)PHOSPHORYLOXY)-N,N,N-TRIMETHYLETHANAMINIUM, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | 著者 | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | 登録日 | 2005-07-26 | | 公開日 | 2005-10-25 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
2AF9
 
 | | Crystal Structure analysis of GM2-Activator protein complexed with phosphatidylcholine | | 分子名称: | Ganglioside GM2 activator, ISOPROPYL ALCOHOL, LAURIC ACID, ... | | 著者 | Wright, C.S, Mi, L.Z, Lee, S, Rastinejad, F. | | 登録日 | 2005-07-25 | | 公開日 | 2005-10-25 | | 最終更新日 | 2024-11-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure Analysis of Phosphatidylcholine-GM2-Activator Product Complexes: Evidence for Hydrolase Activity.
Biochemistry, 44, 2005
|
|
1KN1
 
 | | Crystal structure of allophycocyanin | | 分子名称: | Allophycocyanin, PHYCOCYANOBILIN | | 著者 | Liang, D.C, Liu, J.Y, Jiang, T, Zhang, J.P, Chang, W.R. | | 登録日 | 2001-12-18 | | 公開日 | 2002-12-18 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of Allophycocyanin from red algae Porphyra yezoensis at 2.2 A resolution
J.BIOL.CHEM., 274, 1999
|
|
3O2C
 
 | |
3O18
 
 | |
4I9J
 
 | | Structure of the N254Y/H258Y mutant of the phosphatidylinositol-specific phospholipase C from S. aureus bound to diC4PC | | 分子名称: | (4S,7R)-7-(heptanoyloxy)-4-hydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphahexadecan-1-aminium 4-oxide, 1-phosphatidylinositol phosphodiesterase, ACETATE ION | | 著者 | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | 登録日 | 2012-12-05 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
4I9T
 
 | | Structure of the H258Y mutant of the phosphatidylinositol-specific phospholipase C from Staphylococcus aureus | | 分子名称: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase, SULFATE ION, ... | | 著者 | Goldstein, R.I, Cheng, J, Stec, B, Gershenson, A, Roberts, M.F. | | 登録日 | 2012-12-05 | | 公開日 | 2013-04-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | The cation-pi box is a specific phosphatidylcholine membrane targeting motif.
J.Biol.Chem., 288, 2013
|
|
3EA3
 
 | | Crystal Structure of the Y246S/Y247S/Y248S/Y251S Mutant of Phosphatidylinositol-Specific Phospholipase C from Bacillus Thuringiensis | | 分子名称: | 1-phosphatidylinositol phosphodiesterase, MANGANESE (II) ION | | 著者 | Shi, X, Shao, C, Zhang, X, Zambonelli, C, Redfied, A.G, Head, J.F, Seaton, B.A, Roberts, M.F. | | 登録日 | 2008-08-24 | | 公開日 | 2009-04-14 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Modulation of Bacillus thuringiensis Phosphatidylinositol-specific Phospholipase C Activity by Mutations in the Putative Dimerization Interface.
J.Biol.Chem., 284, 2009
|
|
8USD
 
 | |
